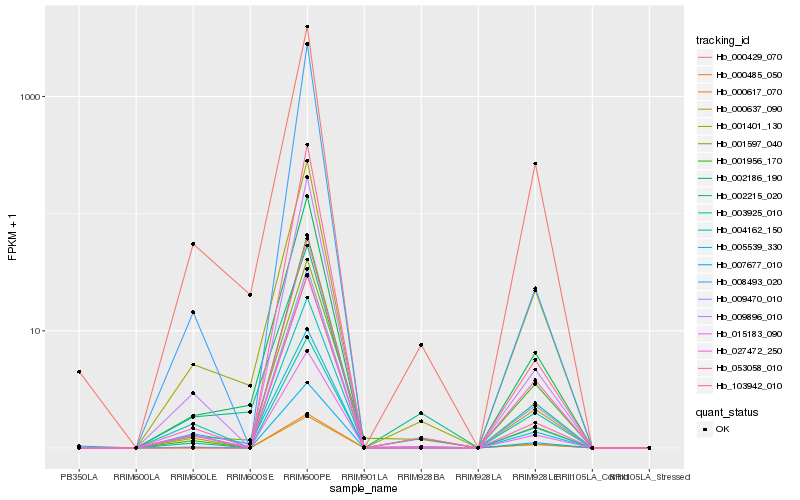
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009470_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000637_090 |
0.0564039574 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 3 |
Hb_007677_010 |
0.0604555391 |
- |
- |
PREDICTED: uncharacterized protein LOC105641028 [Jatropha curcas] |
| 4 |
Hb_001956_170 |
0.065770261 |
- |
- |
hypothetical protein POPTR_0001s24950g [Populus trichocarpa] |
| 5 |
Hb_002186_190 |
0.0761520772 |
- |
- |
- |
| 6 |
Hb_009896_010 |
0.0770250399 |
- |
- |
ABC transporter G family member 15 [Morus notabilis] |
| 7 |
Hb_003925_010 |
0.0778284349 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 8 |
Hb_053058_010 |
0.0786778361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000429_070 |
0.0811999285 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 10 |
Hb_000485_050 |
0.0930935084 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 11 |
Hb_001401_130 |
0.093254223 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 12 |
Hb_004162_150 |
0.096341514 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 13 |
Hb_001597_040 |
0.0972588489 |
- |
- |
- |
| 14 |
Hb_000617_070 |
0.0984400675 |
- |
- |
hypothetical protein POPTR_0005s02420g [Populus trichocarpa] |
| 15 |
Hb_027472_250 |
0.1059688806 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 16 |
Hb_008493_020 |
0.1062715937 |
- |
- |
linamarase [Manihot esculenta] |
| 17 |
Hb_005539_330 |
0.1098260596 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 18 |
Hb_103942_010 |
0.1109754544 |
- |
- |
rab5A-like protein [Trichinella spiralis] |
| 19 |
Hb_002215_020 |
0.1131270951 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 20 |
Hb_015183_090 |
0.1131502916 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |