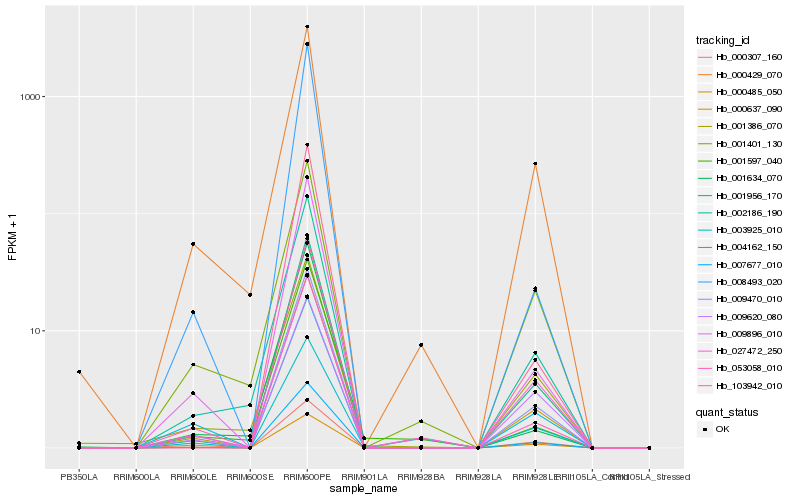
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_090 |
0.0 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 2 |
Hb_007677_010 |
0.0180815986 |
- |
- |
PREDICTED: uncharacterized protein LOC105641028 [Jatropha curcas] |
| 3 |
Hb_001956_170 |
0.0299710404 |
- |
- |
hypothetical protein POPTR_0001s24950g [Populus trichocarpa] |
| 4 |
Hb_009896_010 |
0.0534909446 |
- |
- |
ABC transporter G family member 15 [Morus notabilis] |
| 5 |
Hb_003925_010 |
0.0553971244 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 6 |
Hb_009470_010 |
0.0564039574 |
- |
- |
- |
| 7 |
Hb_002186_190 |
0.0721585749 |
- |
- |
- |
| 8 |
Hb_000485_050 |
0.0759153469 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 9 |
Hb_000429_070 |
0.0802339321 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 10 |
Hb_004162_150 |
0.0820920986 |
- |
- |
hypothetical protein POPTR_0009s00870g [Populus trichocarpa] |
| 11 |
Hb_053058_010 |
0.0879098806 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008493_020 |
0.0896772128 |
- |
- |
linamarase [Manihot esculenta] |
| 13 |
Hb_027472_250 |
0.0902706463 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 14 |
Hb_103942_010 |
0.0935414191 |
- |
- |
rab5A-like protein [Trichinella spiralis] |
| 15 |
Hb_001401_130 |
0.0964450314 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 16 |
Hb_009620_080 |
0.1024720634 |
- |
- |
hypothetical protein POPTR_0007s08150g [Populus trichocarpa] |
| 17 |
Hb_001634_070 |
0.1041981695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000307_160 |
0.1044547794 |
- |
- |
PREDICTED: uncharacterized protein At1g76070 [Jatropha curcas] |
| 19 |
Hb_001386_070 |
0.1044904415 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 20 |
Hb_001597_040 |
0.1076407376 |
- |
- |
- |