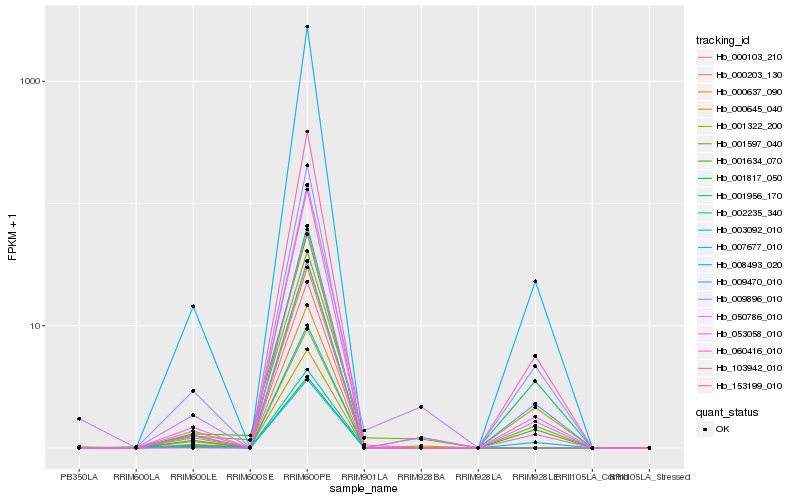
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053058_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008493_020 |
0.0468785502 |
- |
- |
linamarase [Manihot esculenta] |
| 3 |
Hb_009896_010 |
0.0497618784 |
- |
- |
ABC transporter G family member 15 [Morus notabilis] |
| 4 |
Hb_001634_070 |
0.0689393918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001597_040 |
0.0690439604 |
- |
- |
- |
| 6 |
Hb_050786_010 |
0.0721296752 |
- |
- |
hypothetical protein MIMGU_mgv1a014824mg [Erythranthe guttata] |
| 7 |
Hb_060416_010 |
0.0738096177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_103942_010 |
0.0752974966 |
- |
- |
rab5A-like protein [Trichinella spiralis] |
| 9 |
Hb_009470_010 |
0.0786778361 |
- |
- |
- |
| 10 |
Hb_000103_210 |
0.0839808398 |
- |
- |
PREDICTED: protein lin-12-like isoform X1 [Populus euphratica] |
| 11 |
Hb_001322_200 |
0.0840674418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000203_130 |
0.0842244411 |
- |
- |
- |
| 13 |
Hb_000645_040 |
0.0857591505 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001817_050 |
0.0861130004 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 15 |
Hb_002235_340 |
0.0868210016 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000637_090 |
0.0879098806 |
- |
- |
hypothetical protein JCGZ_05877 [Jatropha curcas] |
| 17 |
Hb_153199_010 |
0.0909556694 |
- |
- |
- |
| 18 |
Hb_001956_170 |
0.0936047405 |
- |
- |
hypothetical protein POPTR_0001s24950g [Populus trichocarpa] |
| 19 |
Hb_003092_010 |
0.0936837653 |
- |
- |
hypothetical protein POPTR_0025s00750g [Populus trichocarpa] |
| 20 |
Hb_007677_010 |
0.0948210764 |
- |
- |
PREDICTED: uncharacterized protein LOC105641028 [Jatropha curcas] |