| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
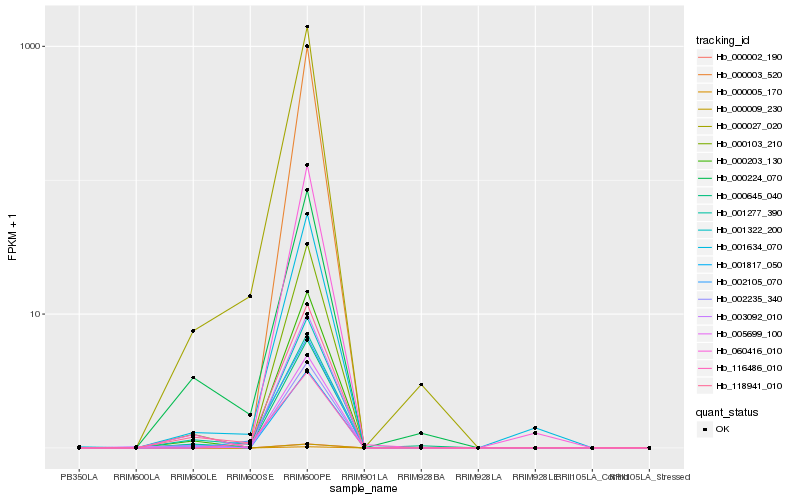
Hb_000027_020 |
0.0 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 2 |
Hb_005699_100 |
0.0589075863 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 3 |
Hb_118941_010 |
0.0626447644 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 4 |
Hb_000645_040 |
0.0638400533 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002105_070 |
0.0714734944 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001634_070 |
0.071684113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001322_200 |
0.0728617956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000103_210 |
0.0739407668 |
- |
- |
PREDICTED: protein lin-12-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000203_130 |
0.0754639804 |
- |
- |
- |
| 10 |
Hb_001277_390 |
0.0772808201 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 11 |
Hb_003092_010 |
0.0796782035 |
- |
- |
hypothetical protein POPTR_0025s00750g [Populus trichocarpa] |
| 12 |
Hb_001817_050 |
0.0802445242 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 13 |
Hb_002235_340 |
0.081605618 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000224_070 |
0.0826458013 |
- |
- |
- |
| 15 |
Hb_060416_010 |
0.0827967526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_116486_010 |
0.0845469984 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 17 |
Hb_000002_190 |
0.0861558274 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 18 |
Hb_000003_520 |
0.0861558274 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 19 |
Hb_000005_170 |
0.0861558274 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR [Jatropha curcas] |
| 20 |
Hb_000009_230 |
0.0861558274 |
- |
- |
hypothetical protein POPTR_0007s12290g [Populus trichocarpa] |