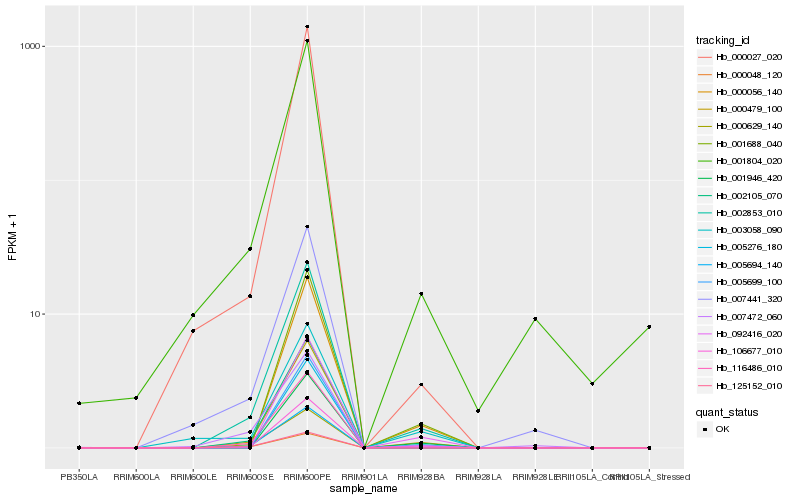
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002853_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000479_100 |
0.0259925538 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 3 |
Hb_106677_010 |
0.0417531773 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 4 |
Hb_116486_010 |
0.0901909643 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 5 |
Hb_005694_140 |
0.0904846126 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002105_070 |
0.0919049962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000629_140 |
0.1074595956 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 8 |
Hb_000027_020 |
0.1075574294 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 9 |
Hb_005699_100 |
0.1117776743 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 10 |
Hb_001804_020 |
0.1139482671 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 11 |
Hb_005276_180 |
0.1194596129 |
- |
- |
hypothetical protein POPTR_0005s23040g [Populus trichocarpa] |
| 12 |
Hb_007472_060 |
0.1216358043 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001688_040 |
0.1217341861 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000048_120 |
0.1218316765 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 15 |
Hb_000056_140 |
0.1224233205 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE6 [Vitis vinifera] |
| 16 |
Hb_125152_010 |
0.1227099803 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 17 |
Hb_003058_090 |
0.1231076378 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 18 |
Hb_007441_320 |
0.1233431169 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 19 |
Hb_001946_420 |
0.1268934488 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_092416_020 |
0.1281154637 |
- |
- |
protein with unknown function [Ricinus communis] |