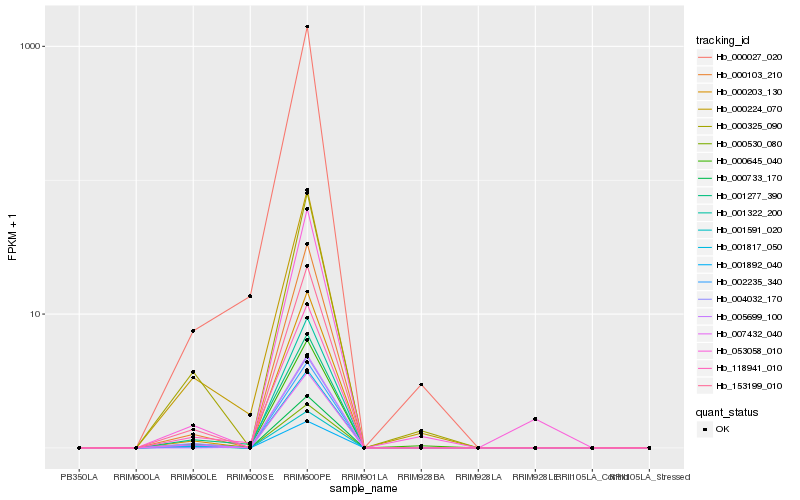
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000645_040 |
0.0 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000224_070 |
0.0616576826 |
- |
- |
- |
| 3 |
Hb_000027_020 |
0.0638400533 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 4 |
Hb_001817_050 |
0.0691039541 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 5 |
Hb_002235_340 |
0.0691263287 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 6 |
Hb_118941_010 |
0.0698388001 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 7 |
Hb_000203_130 |
0.0704261939 |
- |
- |
- |
| 8 |
Hb_153199_010 |
0.0707212865 |
- |
- |
- |
| 9 |
Hb_000325_090 |
0.0711819134 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 10 |
Hb_000103_210 |
0.0716734801 |
- |
- |
PREDICTED: protein lin-12-like isoform X1 [Populus euphratica] |
| 11 |
Hb_001322_200 |
0.0731488391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007432_040 |
0.0738760767 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 13 |
Hb_004032_170 |
0.0741450546 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 14 |
Hb_000530_080 |
0.0748223065 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2-like [Jatropha curcas] |
| 15 |
Hb_000733_170 |
0.0761598873 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 16 |
Hb_001591_020 |
0.078783065 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 17 |
Hb_001277_390 |
0.0802201972 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 18 |
Hb_001892_040 |
0.0813465377 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 19 |
Hb_005699_100 |
0.0833708033 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 20 |
Hb_053058_010 |
0.0857591505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |