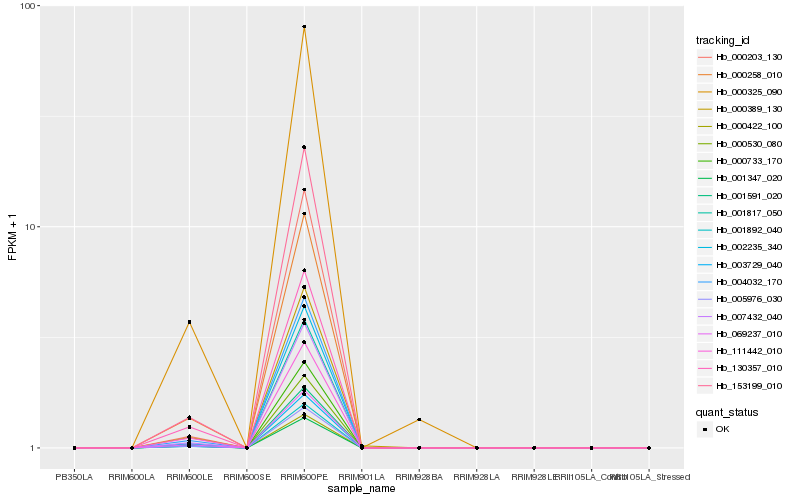
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001892_040 |
0.0 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 2 |
Hb_001591_020 |
0.0051655208 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 3 |
Hb_000733_170 |
0.0110588264 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 4 |
Hb_005976_030 |
0.0128791624 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 5 |
Hb_000530_080 |
0.0144178933 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2-like [Jatropha curcas] |
| 6 |
Hb_004032_170 |
0.0162478802 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG7 [Jatropha curcas] |
| 7 |
Hb_007432_040 |
0.017004309 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 8 |
Hb_000258_010 |
0.0179290414 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 9 |
Hb_153199_010 |
0.0281062882 |
- |
- |
- |
| 10 |
Hb_069237_010 |
0.0302195387 |
- |
- |
- |
| 11 |
Hb_003729_040 |
0.0360564654 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_130357_010 |
0.0402523883 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_002235_340 |
0.0410588067 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001817_050 |
0.0440131059 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 15 |
Hb_000325_090 |
0.0486391233 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 16 |
Hb_000389_130 |
0.0500140238 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 17 |
Hb_001347_020 |
0.0507622033 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Populus euphratica] |
| 18 |
Hb_000422_100 |
0.0520211867 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 19 |
Hb_111442_010 |
0.0548303712 |
desease resistance |
Gene Name: AAA_16 |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 20 |
Hb_000203_130 |
0.0563263638 |
- |
- |
- |