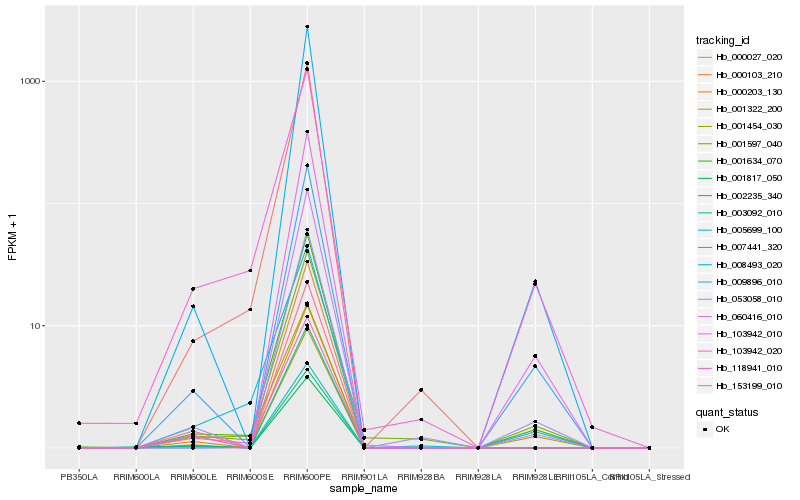
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001634_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008493_020 |
0.0515025427 |
- |
- |
linamarase [Manihot esculenta] |
| 3 |
Hb_060416_010 |
0.0648059244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009896_010 |
0.06750992 |
- |
- |
ABC transporter G family member 15 [Morus notabilis] |
| 5 |
Hb_053058_010 |
0.0689393918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001597_040 |
0.0706966613 |
- |
- |
- |
| 7 |
Hb_000027_020 |
0.071684113 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 8 |
Hb_103942_010 |
0.0755960876 |
- |
- |
rab5A-like protein [Trichinella spiralis] |
| 9 |
Hb_001454_030 |
0.0763399051 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 10 |
Hb_001322_200 |
0.0799696431 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000103_210 |
0.0806407836 |
- |
- |
PREDICTED: protein lin-12-like isoform X1 [Populus euphratica] |
| 12 |
Hb_000203_130 |
0.0817080728 |
- |
- |
- |
| 13 |
Hb_118941_010 |
0.0834523113 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 14 |
Hb_001817_050 |
0.0854315145 |
transcription factor |
TF Family: C2H2 |
PREDICTED: transcriptional regulator SUPERMAN [Jatropha curcas] |
| 15 |
Hb_002235_340 |
0.0865491022 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_103942_020 |
0.0867278273 |
- |
- |
unknown [Zea mays] |
| 17 |
Hb_003092_010 |
0.0873664899 |
- |
- |
hypothetical protein POPTR_0025s00750g [Populus trichocarpa] |
| 18 |
Hb_005699_100 |
0.0901857031 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 19 |
Hb_007441_320 |
0.0920402753 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 20 |
Hb_153199_010 |
0.0923094304 |
- |
- |
- |