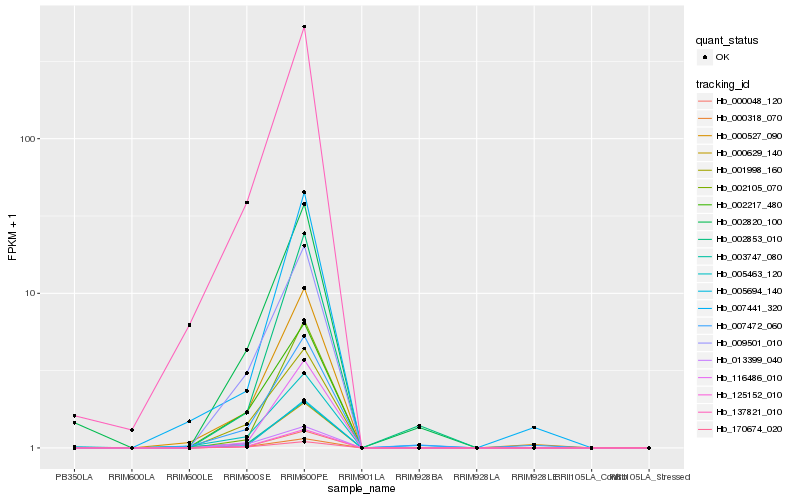
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000629_140 |
0.0 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 2 |
Hb_000048_120 |
0.0239751014 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 3 |
Hb_125152_010 |
0.0253077714 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 4 |
Hb_005694_140 |
0.0513881881 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_116486_010 |
0.0575403794 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 6 |
Hb_009501_010 |
0.0633570938 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_137821_010 |
0.0754834327 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 8 |
Hb_002105_070 |
0.0760892774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000527_090 |
0.0818055522 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 10 |
Hb_001998_160 |
0.0835579088 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |
| 11 |
Hb_002217_480 |
0.096397071 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 12 |
Hb_000318_070 |
0.0997269193 |
- |
- |
PREDICTED: peroxidase 10-like [Jatropha curcas] |
| 13 |
Hb_003747_080 |
0.1044983379 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_170674_020 |
0.1054105449 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |
| 15 |
Hb_002853_010 |
0.1074595956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002820_100 |
0.1090575495 |
- |
- |
hypothetical protein OsI_33031 [Oryza sativa Indica Group] |
| 17 |
Hb_005463_120 |
0.1096743293 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 18 |
Hb_007441_320 |
0.1131399101 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 19 |
Hb_013399_040 |
0.1140141586 |
- |
- |
hypothetical protein JCGZ_24713 [Jatropha curcas] |
| 20 |
Hb_007472_060 |
0.114469361 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |