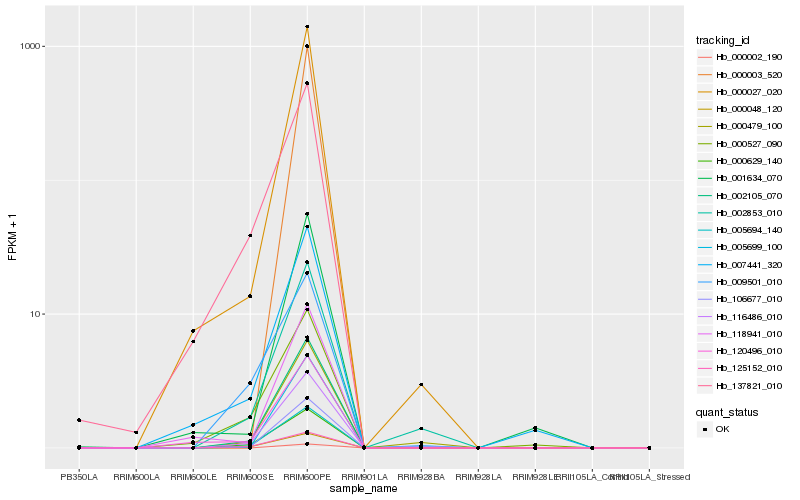
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116486_010 |
0.0 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 2 |
Hb_005694_140 |
0.0062323761 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_002105_070 |
0.0189488324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000629_140 |
0.0575403794 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 5 |
Hb_000048_120 |
0.0811512682 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 6 |
Hb_125152_010 |
0.0824593937 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 7 |
Hb_000027_020 |
0.0845469984 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 8 |
Hb_002853_010 |
0.0901909643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007441_320 |
0.0979889215 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 10 |
Hb_000479_100 |
0.0983846582 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 11 |
Hb_005699_100 |
0.1010087054 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 12 |
Hb_137821_010 |
0.104790539 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 13 |
Hb_000527_090 |
0.1091781692 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_120496_010 |
0.1114479905 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 15 |
Hb_106677_010 |
0.1125679673 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 16 |
Hb_118941_010 |
0.1172005034 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 17 |
Hb_001634_070 |
0.1173546046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_009501_010 |
0.1196638525 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_000002_190 |
0.1214063302 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 20 |
Hb_000003_520 |
0.1214063302 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |