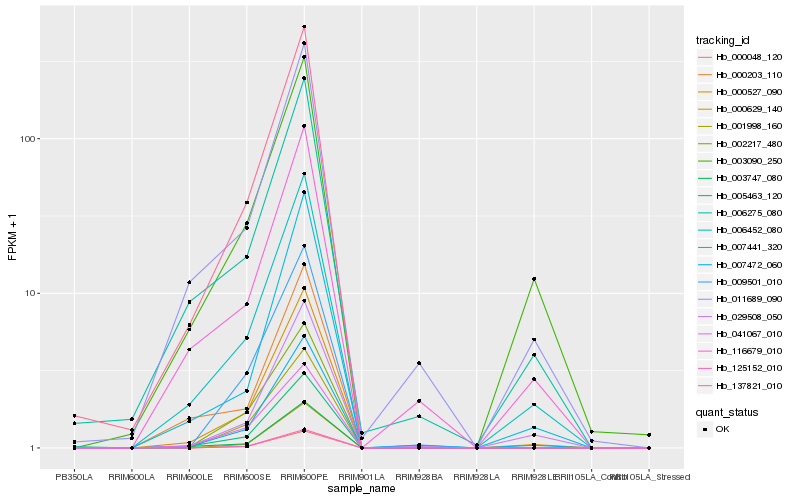
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000527_090 |
0.0 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 2 |
Hb_006452_080 |
0.0487868563 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 3 |
Hb_137821_010 |
0.0573484772 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 4 |
Hb_003747_080 |
0.0708632243 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 5 |
Hb_007441_320 |
0.0785016511 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 6 |
Hb_007472_060 |
0.0797293573 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000048_120 |
0.0802645636 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g37970 [Jatropha curcas] |
| 8 |
Hb_125152_010 |
0.0803870165 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 9 |
Hb_000629_140 |
0.0818055522 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 10 |
Hb_011689_090 |
0.0839992179 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 11 |
Hb_005463_120 |
0.0863998032 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 12 |
Hb_029508_050 |
0.0890486091 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 13 |
Hb_009501_010 |
0.0923080627 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_003090_250 |
0.0946526045 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_006275_080 |
0.094874014 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 16 |
Hb_116679_010 |
0.0950106755 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 17 |
Hb_002217_480 |
0.0950261085 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 18 |
Hb_041067_010 |
0.0985424565 |
- |
- |
hypothetical protein JCGZ_00780 [Jatropha curcas] |
| 19 |
Hb_000203_110 |
0.0998855552 |
- |
- |
- |
| 20 |
Hb_001998_160 |
0.1038883899 |
- |
- |
PREDICTED: origin recognition complex subunit 1-like [Populus euphratica] |