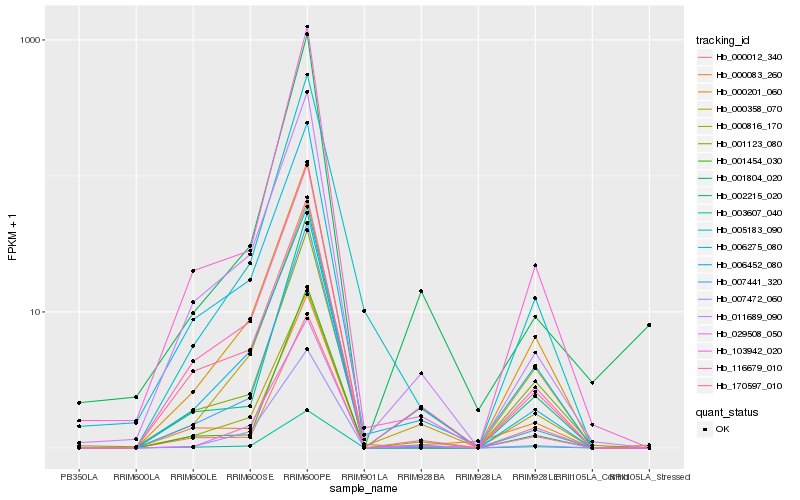
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003607_040 |
0.0 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002215_020 |
0.0573676629 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 3 |
Hb_116679_010 |
0.0626233924 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 4 |
Hb_000012_340 |
0.0711198723 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 5 |
Hb_000358_070 |
0.0713160621 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 6 |
Hb_000816_170 |
0.0735652087 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 7 |
Hb_029508_050 |
0.0741263779 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_103942_020 |
0.0741266492 |
- |
- |
unknown [Zea mays] |
| 9 |
Hb_007441_320 |
0.0744755039 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 10 |
Hb_011689_090 |
0.0750369689 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 11 |
Hb_170597_010 |
0.0834674015 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 12 |
Hb_001454_030 |
0.0838488537 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 13 |
Hb_000201_060 |
0.0886767086 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X2 [Populus euphratica] |
| 14 |
Hb_006452_080 |
0.0889962677 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 15 |
Hb_007472_060 |
0.0923452501 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000083_260 |
0.0948900362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006275_080 |
0.0967368479 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 18 |
Hb_001804_020 |
0.0981306622 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 19 |
Hb_005183_090 |
0.1000836431 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 20 |
Hb_001123_080 |
0.1011651039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |