| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
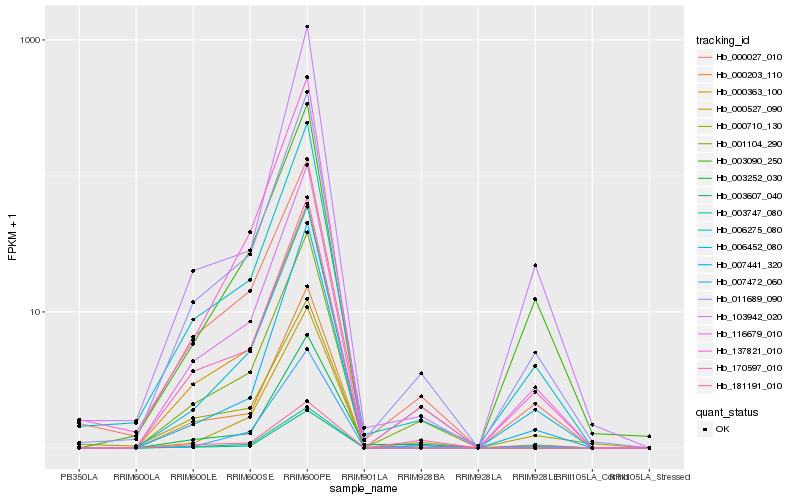
Hb_011689_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 2 |
Hb_116679_010 |
0.0323324629 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 3 |
Hb_006275_080 |
0.0451462386 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 4 |
Hb_170597_010 |
0.0619642989 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 5 |
Hb_006452_080 |
0.0699501337 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 6 |
Hb_003607_040 |
0.0750369689 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000027_010 |
0.0768943087 |
- |
- |
- |
| 8 |
Hb_001104_290 |
0.0808261277 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 9 |
Hb_007472_060 |
0.080838713 |
- |
- |
PREDICTED: ABC transporter G family member 14-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000363_100 |
0.0826251172 |
- |
- |
- |
| 11 |
Hb_000527_090 |
0.0839992179 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_007441_320 |
0.086104658 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 13 |
Hb_003747_080 |
0.0885495099 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_000203_110 |
0.0933185892 |
- |
- |
- |
| 15 |
Hb_003252_030 |
0.0978390959 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 16 |
Hb_103942_020 |
0.0986076743 |
- |
- |
unknown [Zea mays] |
| 17 |
Hb_003090_250 |
0.0990961104 |
- |
- |
Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_181191_010 |
0.0993629646 |
- |
- |
kinase, putative [Ricinus communis] |
| 19 |
Hb_000710_130 |
0.1015117951 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 20 |
Hb_137821_010 |
0.1023303039 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |