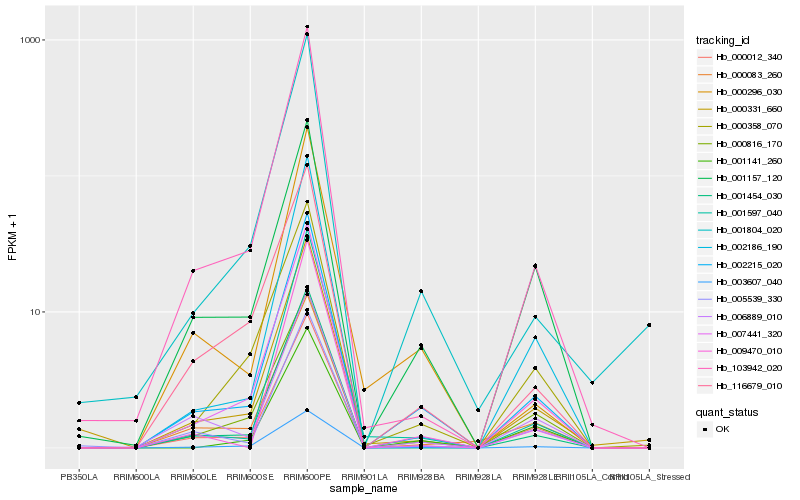
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002215_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 2 |
Hb_003607_040 |
0.0573676629 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000012_340 |
0.07659168 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 4 |
Hb_103942_020 |
0.0940583646 |
- |
- |
unknown [Zea mays] |
| 5 |
Hb_002186_190 |
0.0966536792 |
- |
- |
- |
| 6 |
Hb_001454_030 |
0.0976531644 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 7 |
Hb_000816_170 |
0.0989437274 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 8 |
Hb_005539_330 |
0.1018594007 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 9 |
Hb_000296_030 |
0.1040783108 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 10 |
Hb_001804_020 |
0.1045665585 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 11 |
Hb_116679_010 |
0.1053949554 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 12 |
Hb_001157_120 |
0.1058499206 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 13 |
Hb_006889_010 |
0.1058820108 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 14 |
Hb_000083_260 |
0.1061237207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007441_320 |
0.1065614635 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 16 |
Hb_000358_070 |
0.1083337547 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 17 |
Hb_000331_660 |
0.1098847929 |
- |
- |
PREDICTED: vignain [Jatropha curcas] |
| 18 |
Hb_001597_040 |
0.1103582771 |
- |
- |
- |
| 19 |
Hb_009470_010 |
0.1131270951 |
- |
- |
- |
| 20 |
Hb_001141_260 |
0.1143101862 |
- |
- |
Uncharacterized protein TCM_020647 [Theobroma cacao] |