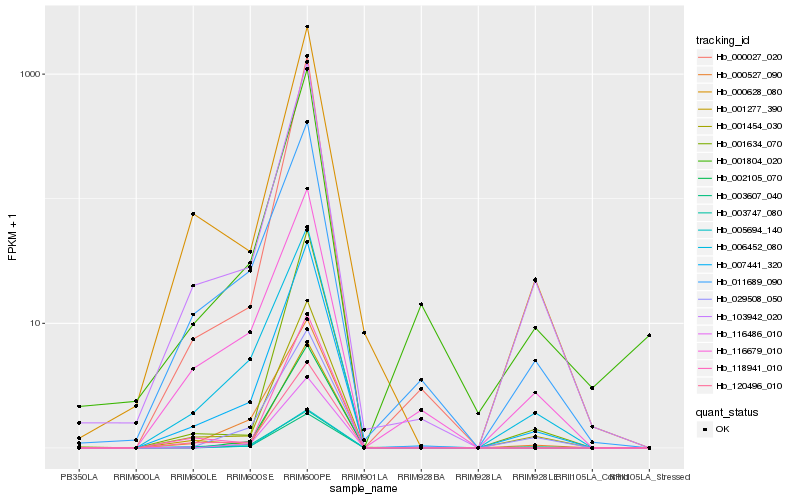
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_320 |
0.0 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 2 |
Hb_103942_020 |
0.0507055629 |
- |
- |
unknown [Zea mays] |
| 3 |
Hb_001454_030 |
0.0544971446 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 3 [Jatropha curcas] |
| 4 |
Hb_003607_040 |
0.0744755039 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000527_090 |
0.0785016511 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_120496_010 |
0.0799062029 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 7 |
Hb_006452_080 |
0.0819097576 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 8 |
Hb_000628_080 |
0.0859006509 |
- |
- |
hypothetical protein JCGZ_04283 [Jatropha curcas] |
| 9 |
Hb_011689_090 |
0.086104658 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 10 |
Hb_001634_070 |
0.0920402753 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_029508_050 |
0.0920528818 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like isoform X2 [Musa acuminata subsp. malaccensis] |
| 12 |
Hb_116679_010 |
0.0935959745 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 13 |
Hb_000027_020 |
0.0950697952 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 14 |
Hb_001804_020 |
0.0972727924 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 15 |
Hb_003747_080 |
0.0972801944 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_116486_010 |
0.0979889215 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 17 |
Hb_118941_010 |
0.0981095258 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 18 |
Hb_005694_140 |
0.0981418531 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_001277_390 |
0.0983036534 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 5 [Jatropha curcas] |
| 20 |
Hb_002105_070 |
0.0999164435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |