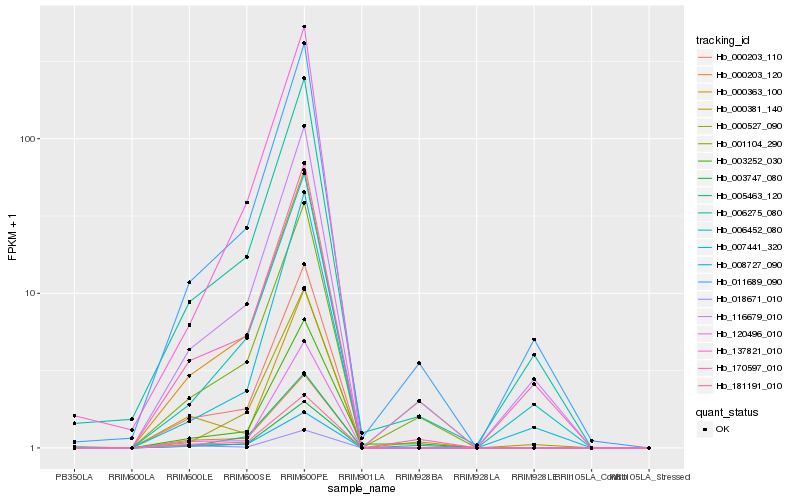
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000203_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_181191_010 |
0.0326356926 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_003747_080 |
0.0374332178 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_000203_120 |
0.0514505063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_008727_090 |
0.0528576671 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 6 |
Hb_120496_010 |
0.0578205656 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 7 |
Hb_137821_010 |
0.0865432357 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |
| 8 |
Hb_001104_290 |
0.0893730599 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 9 |
Hb_000363_100 |
0.0905977399 |
- |
- |
- |
| 10 |
Hb_011689_090 |
0.0933185892 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 11 |
Hb_006275_080 |
0.0975206046 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 12 |
Hb_000527_090 |
0.0998855552 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 13 |
Hb_000381_140 |
0.1027690267 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 14 |
Hb_003252_030 |
0.1030644031 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 15 |
Hb_005463_120 |
0.1034317457 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 16 |
Hb_006452_080 |
0.1054407194 |
- |
- |
Nodulin, putative [Ricinus communis] |
| 17 |
Hb_116679_010 |
0.106324025 |
- |
- |
PREDICTED: transcription factor bHLH110 isoform X2 [Vitis vinifera] |
| 18 |
Hb_170597_010 |
0.1068582304 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 19 |
Hb_007441_320 |
0.1084352478 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 20 |
Hb_018671_010 |
0.1084919994 |
- |
- |
- |