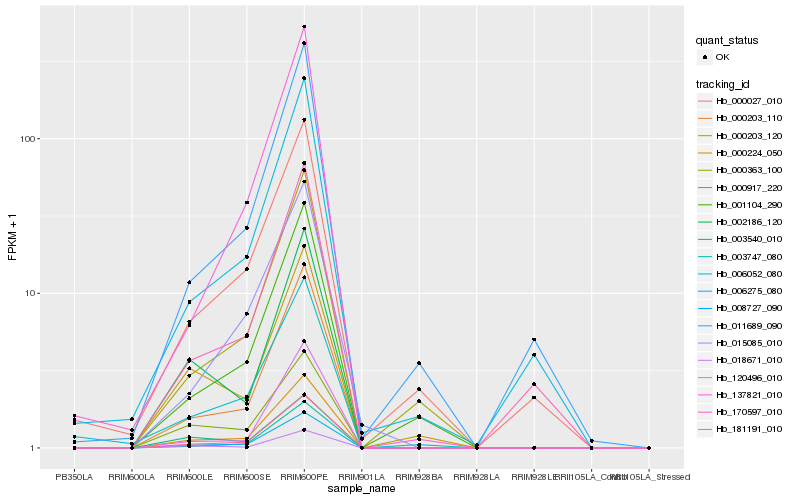
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008727_090 |
0.0 |
- |
- |
PREDICTED: 17.3 kDa class I heat shock protein-like [Jatropha curcas] |
| 2 |
Hb_000203_120 |
0.005943904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_181191_010 |
0.0243982082 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000203_110 |
0.0528576671 |
- |
- |
- |
| 5 |
Hb_003747_080 |
0.0752625303 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 6 |
Hb_000917_220 |
0.0830382189 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 7 |
Hb_018671_010 |
0.0952654898 |
- |
- |
- |
| 8 |
Hb_002186_120 |
0.0985877476 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 9 |
Hb_003540_010 |
0.099511204 |
- |
- |
- |
| 10 |
Hb_000363_100 |
0.1012552772 |
- |
- |
- |
| 11 |
Hb_001104_290 |
0.1025936661 |
transcription factor |
TF Family: ERF |
Dehydration-responsive element-binding protein 1B, putative [Theobroma cacao] |
| 12 |
Hb_015085_010 |
0.1057192114 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 13 |
Hb_000224_050 |
0.1087006239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006052_080 |
0.1088031645 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH140-like [Populus euphratica] |
| 15 |
Hb_120496_010 |
0.1089783521 |
- |
- |
PREDICTED: CSC1-like protein At1g32090 [Cicer arietinum] |
| 16 |
Hb_006275_080 |
0.1094845505 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 17 |
Hb_011689_090 |
0.1124357137 |
- |
- |
PREDICTED: uncharacterized protein LOC105631865 [Jatropha curcas] |
| 18 |
Hb_000027_010 |
0.1130310582 |
- |
- |
- |
| 19 |
Hb_170597_010 |
0.1151671663 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 20 |
Hb_137821_010 |
0.1163276257 |
- |
- |
hypothetical protein CISIN_1g034189mg [Citrus sinensis] |