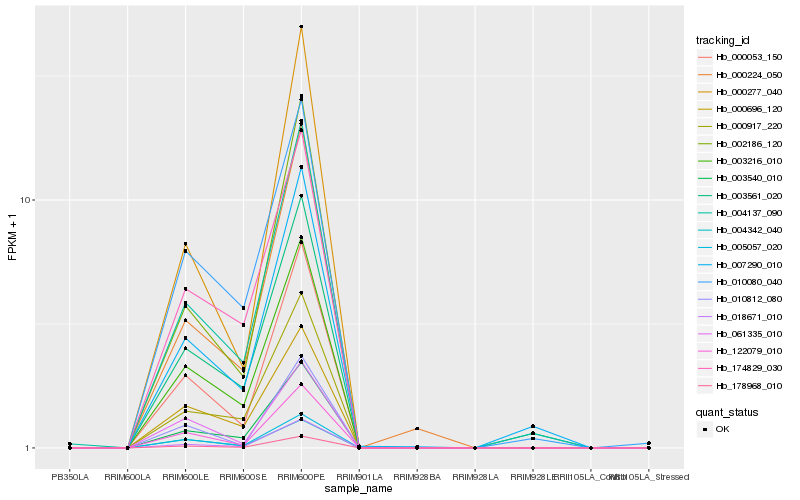
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178968_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 2 |
Hb_000053_150 |
0.0250980633 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 3 |
Hb_003540_010 |
0.0450543036 |
- |
- |
- |
| 4 |
Hb_018671_010 |
0.0475756476 |
- |
- |
- |
| 5 |
Hb_003216_010 |
0.0476894264 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 6 |
Hb_122079_010 |
0.0500453217 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 7 |
Hb_002186_120 |
0.0509094037 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 8 |
Hb_005057_020 |
0.0540554014 |
- |
- |
PREDICTED: uncharacterized protein LOC105631610 [Jatropha curcas] |
| 9 |
Hb_004137_090 |
0.0646944531 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 10 |
Hb_000277_040 |
0.0678046631 |
- |
- |
- |
| 11 |
Hb_000917_220 |
0.0697522739 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 12 |
Hb_000224_050 |
0.0753069585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010812_080 |
0.0833365968 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 14 |
Hb_000696_120 |
0.0873312324 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 15 |
Hb_174829_030 |
0.0879281569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003561_020 |
0.0888037298 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 17 |
Hb_061335_010 |
0.0910258127 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 18 |
Hb_007290_010 |
0.0916389486 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_004342_040 |
0.0919259901 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 20 |
Hb_010080_040 |
0.0982876734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |