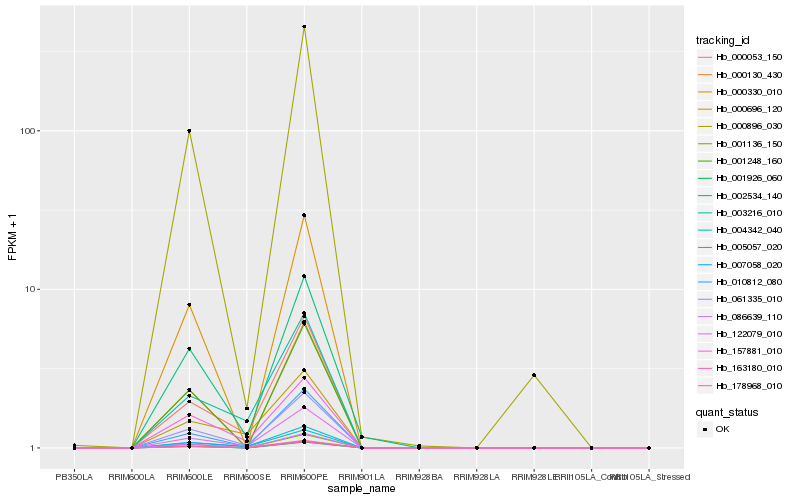
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061335_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 2 |
Hb_122079_010 |
0.0478780319 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 3 |
Hb_004342_040 |
0.0530554931 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 4 |
Hb_005057_020 |
0.0614713421 |
- |
- |
PREDICTED: uncharacterized protein LOC105631610 [Jatropha curcas] |
| 5 |
Hb_157881_010 |
0.0682632464 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_000053_150 |
0.0711065752 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 7 |
Hb_010812_080 |
0.0719608541 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 8 |
Hb_002534_140 |
0.0884752279 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |
| 9 |
Hb_178968_010 |
0.0910258127 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 10 |
Hb_003216_010 |
0.0965062364 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 11 |
Hb_000896_030 |
0.1061687827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001136_150 |
0.1089498608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000130_430 |
0.1089594417 |
- |
- |
PREDICTED: UDP-glycosyltransferase 79B6-like [Jatropha curcas] |
| 14 |
Hb_001248_160 |
0.1089733431 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 15 |
Hb_163180_010 |
0.1091328926 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 16 |
Hb_000330_010 |
0.1091525351 |
- |
- |
hypothetical protein POPTR_0005s25020g [Populus trichocarpa] |
| 17 |
Hb_086639_110 |
0.1091620304 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Theobroma cacao] |
| 18 |
Hb_000696_120 |
0.1091953472 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 19 |
Hb_001926_060 |
0.1092028359 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007058_020 |
0.1092740675 |
- |
- |
- |