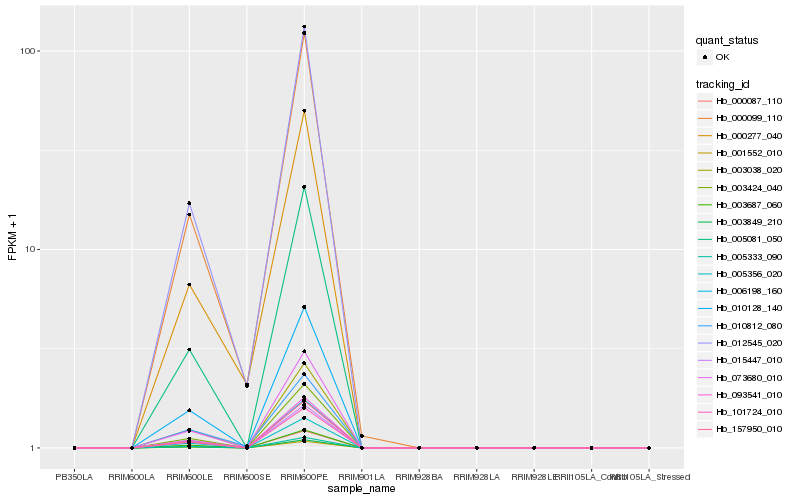
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012545_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 2 |
Hb_000099_110 |
0.0250468053 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 3 |
Hb_000277_040 |
0.0476493738 |
- |
- |
- |
| 4 |
Hb_010812_080 |
0.0547015262 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 5 |
Hb_006198_160 |
0.0595085803 |
- |
- |
hypothetical protein POPTR_0002s24290g [Populus trichocarpa] |
| 6 |
Hb_015447_010 |
0.0595300682 |
- |
- |
PREDICTED: uncharacterized protein LOC105111046 [Populus euphratica] |
| 7 |
Hb_101724_010 |
0.0597864144 |
- |
- |
glutamate receptor family protein [Populus trichocarpa] |
| 8 |
Hb_093541_010 |
0.0598962775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003849_210 |
0.0600341317 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0793640 [Ricinus communis] |
| 10 |
Hb_000087_110 |
0.0601511956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010128_140 |
0.0604228863 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 12 |
Hb_001552_010 |
0.0611194376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005356_020 |
0.0612896568 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 14 |
Hb_005081_050 |
0.0615160173 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor WER-like [Prunus mume] |
| 15 |
Hb_073680_010 |
0.0620107604 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 16 |
Hb_157950_010 |
0.0620425513 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003687_060 |
0.0622794146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003424_040 |
0.0624203662 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 19 |
Hb_003038_020 |
0.0624971344 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_005333_090 |
0.0627325453 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |