| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
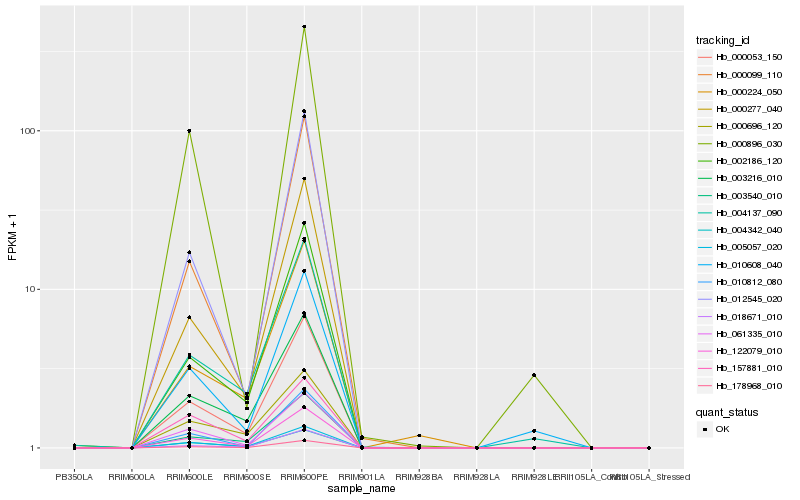
Hb_122079_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 2 |
Hb_000053_150 |
0.0259207877 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 3 |
Hb_010812_080 |
0.045946933 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 4 |
Hb_061335_010 |
0.0478780319 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 5 |
Hb_178968_010 |
0.0500453217 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 6 |
Hb_005057_020 |
0.0536158181 |
- |
- |
PREDICTED: uncharacterized protein LOC105631610 [Jatropha curcas] |
| 7 |
Hb_000277_040 |
0.0730854305 |
- |
- |
- |
| 8 |
Hb_004342_040 |
0.0751421533 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 9 |
Hb_003216_010 |
0.0761890068 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 10 |
Hb_002186_120 |
0.0801507257 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 11 |
Hb_018671_010 |
0.080258597 |
- |
- |
- |
| 12 |
Hb_012545_020 |
0.0907369876 |
- |
- |
PREDICTED: uncharacterized protein LOC105633903 [Jatropha curcas] |
| 13 |
Hb_003540_010 |
0.0917219175 |
- |
- |
- |
| 14 |
Hb_004137_090 |
0.0924860626 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 15 |
Hb_000099_110 |
0.0972746241 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 16 |
Hb_010608_040 |
0.1021197708 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 17 |
Hb_000696_120 |
0.1051786222 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 18 |
Hb_000224_050 |
0.1054107027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_157881_010 |
0.1069830224 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_000896_030 |
0.1098545072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |