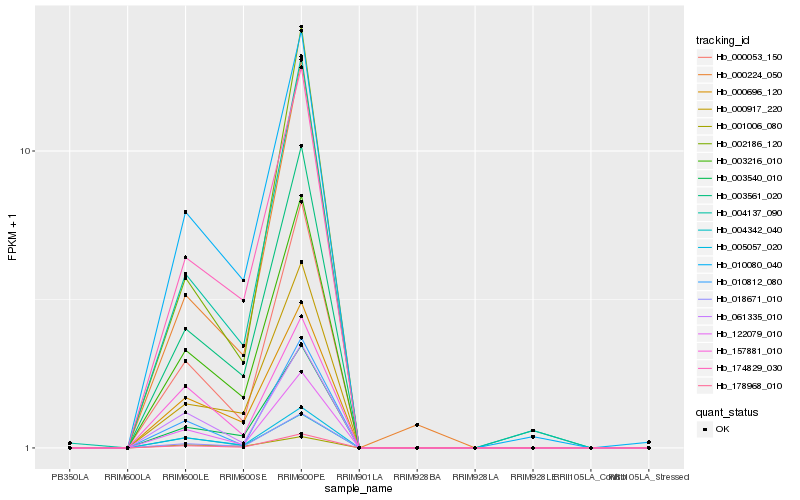
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005057_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631610 [Jatropha curcas] |
| 2 |
Hb_003216_010 |
0.0363804013 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 3 |
Hb_004342_040 |
0.0390721148 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 4 |
Hb_000053_150 |
0.0525328804 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 5 |
Hb_000696_120 |
0.0534655352 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 6 |
Hb_122079_010 |
0.0536158181 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 7 |
Hb_178968_010 |
0.0540554014 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 8 |
Hb_061335_010 |
0.0614713421 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 9 |
Hb_003540_010 |
0.0701429888 |
- |
- |
- |
| 10 |
Hb_010080_040 |
0.0759382458 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_174829_030 |
0.0761678019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_157881_010 |
0.0806635191 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_004137_090 |
0.0875424677 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 14 |
Hb_001006_080 |
0.0888255255 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 15 |
Hb_000917_220 |
0.0949641921 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 16 |
Hb_003561_020 |
0.0952319897 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 17 |
Hb_010812_080 |
0.098267846 |
- |
- |
PREDICTED: methylesterase 17 [Jatropha curcas] |
| 18 |
Hb_018671_010 |
0.1011804306 |
- |
- |
- |
| 19 |
Hb_002186_120 |
0.1040630234 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 20 |
Hb_000224_050 |
0.109745642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |