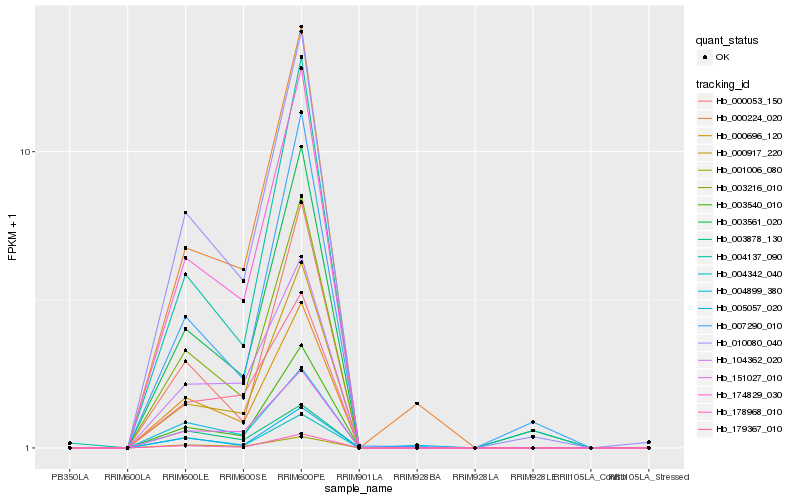
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010080_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000696_120 |
0.0475282437 |
- |
- |
PREDICTED: protein FD [Populus euphratica] |
| 3 |
Hb_174829_030 |
0.0518750143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001006_080 |
0.0606014028 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 5 |
Hb_003216_010 |
0.060614624 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 6 |
Hb_003561_020 |
0.0751972496 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 7 |
Hb_005057_020 |
0.0759382458 |
- |
- |
PREDICTED: uncharacterized protein LOC105631610 [Jatropha curcas] |
| 8 |
Hb_003540_010 |
0.0793294796 |
- |
- |
- |
| 9 |
Hb_151027_010 |
0.0800166971 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 10 |
Hb_000917_220 |
0.0880738268 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 11 |
Hb_004342_040 |
0.0893115756 |
- |
- |
PREDICTED: carotenoid 9,10(9',10')-cleavage dioxygenase 1-like [Jatropha curcas] |
| 12 |
Hb_004137_090 |
0.0910047375 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 13 |
Hb_104362_020 |
0.0915535919 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_178968_010 |
0.0982876734 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 15 |
Hb_003878_130 |
0.1012549614 |
- |
- |
- |
| 16 |
Hb_004899_380 |
0.1078640101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000224_020 |
0.108129103 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 18 |
Hb_179367_010 |
0.1099238767 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 19 |
Hb_000053_150 |
0.1101773918 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 20 |
Hb_007290_010 |
0.1102977054 |
- |
- |
cytochrome P450, putative [Ricinus communis] |