| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
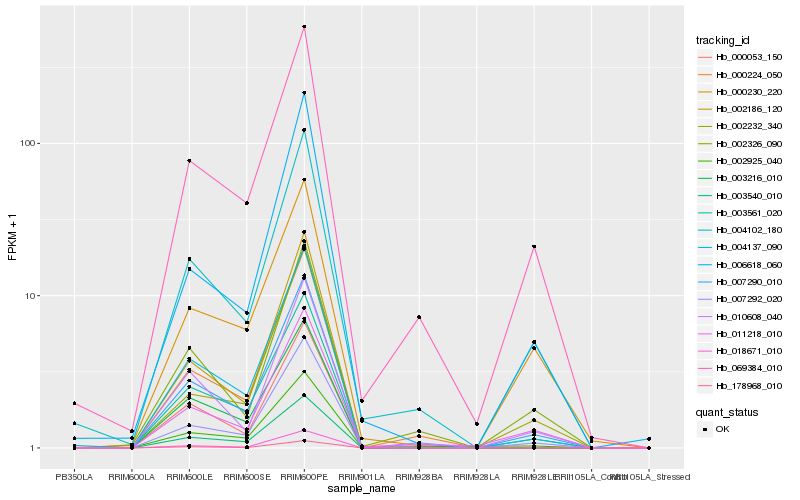
| 1 |
Hb_007290_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_003561_020 |
0.0475160972 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 3 |
Hb_004137_090 |
0.0550447266 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 4 |
Hb_004102_180 |
0.0732668125 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_069384_010 |
0.078467533 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_010608_040 |
0.0795102916 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 7 |
Hb_011218_010 |
0.0906097978 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 8 |
Hb_178968_010 |
0.0916389486 |
- |
- |
hypothetical protein JCGZ_15830 [Jatropha curcas] |
| 9 |
Hb_007292_020 |
0.0962303926 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 10 |
Hb_003540_010 |
0.0966467437 |
- |
- |
- |
| 11 |
Hb_000053_150 |
0.0986456511 |
- |
- |
clavata3/esr-related 12 family protein [Populus trichocarpa] |
| 12 |
Hb_002326_090 |
0.0990182805 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_002232_340 |
0.0993361574 |
- |
- |
Sodium transporter hkt1-like protein, putative [Theobroma cacao] |
| 14 |
Hb_018671_010 |
0.0995321582 |
- |
- |
- |
| 15 |
Hb_002186_120 |
0.1015390627 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X5 [Populus euphratica] |
| 16 |
Hb_000230_220 |
0.1026322651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003216_010 |
0.1028019737 |
- |
- |
glycosyl hydrolase family protein [Populus trichocarpa] |
| 18 |
Hb_006618_060 |
0.1045390127 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 19 |
Hb_002925_040 |
0.1059198843 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 20 |
Hb_000224_050 |
0.1059349137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |