| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
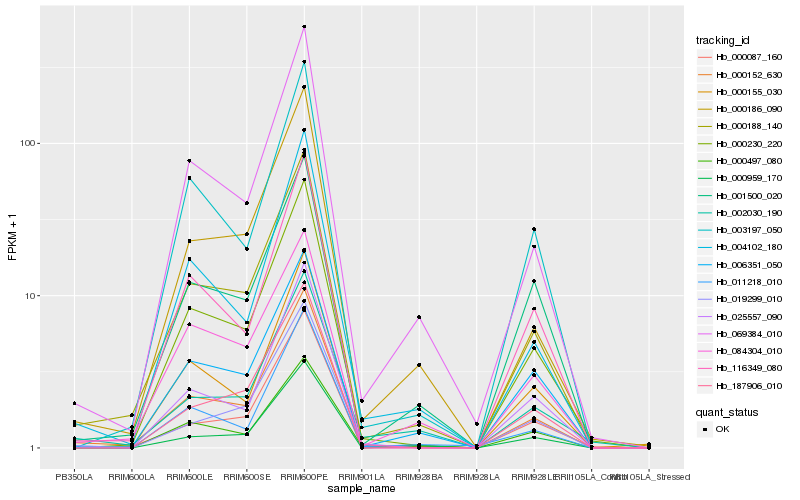
Hb_000152_630 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 [Jatropha curcas] |
| 2 |
Hb_000188_140 |
0.0319212121 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 3 |
Hb_000230_220 |
0.0737213701 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_069384_010 |
0.0802484548 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004102_180 |
0.087997521 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 6 |
Hb_000087_160 |
0.0905703125 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_116349_080 |
0.0923207415 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000186_090 |
0.0930612132 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 9 |
Hb_003197_050 |
0.097132542 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 10 |
Hb_011218_010 |
0.100221554 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 11 |
Hb_006351_050 |
0.1029060747 |
- |
- |
PREDICTED: uncharacterized protein LOC105133646 [Populus euphratica] |
| 12 |
Hb_002030_190 |
0.1057809665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000959_170 |
0.1063362244 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 14 |
Hb_025557_090 |
0.108631806 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 15 |
Hb_019299_010 |
0.1094548434 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_000155_030 |
0.109731157 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 17 |
Hb_000497_080 |
0.1097975512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_084304_010 |
0.1099208592 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X2 [Jatropha curcas] |
| 19 |
Hb_187906_010 |
0.1099782203 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA2-like [Populus euphratica] |
| 20 |
Hb_001500_020 |
0.1119709489 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |