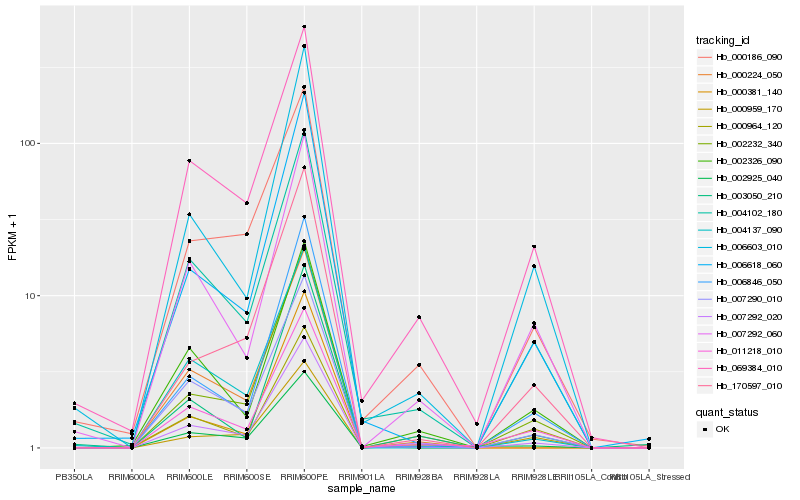
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007292_020 |
0.0 |
- |
- |
Indole-3-acetic acid-amido synthetase GH3.6, putative [Ricinus communis] |
| 2 |
Hb_069384_010 |
0.079609451 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004102_180 |
0.0926213763 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_011218_010 |
0.0944380398 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 5 |
Hb_000186_090 |
0.0951510631 |
- |
- |
ACC oxidase 1 [Hevea brasiliensis] |
| 6 |
Hb_000224_050 |
0.0960114641 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007290_010 |
0.0962303926 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_002326_090 |
0.1002678319 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_002232_340 |
0.1042824662 |
- |
- |
Sodium transporter hkt1-like protein, putative [Theobroma cacao] |
| 10 |
Hb_007292_060 |
0.1047002708 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 11 |
Hb_006603_010 |
0.1048470047 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 12 |
Hb_000964_120 |
0.1053571394 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 13 |
Hb_006846_050 |
0.1064636627 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_006618_060 |
0.1080158985 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 15 |
Hb_002925_040 |
0.1102234224 |
desease resistance |
Gene Name: TIR |
hypothetical protein JCGZ_01545 [Jatropha curcas] |
| 16 |
Hb_003050_210 |
0.1111801081 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 17 |
Hb_000959_170 |
0.1115738068 |
- |
- |
Cell division protein kinase 7, putative [Ricinus communis] |
| 18 |
Hb_000381_140 |
0.1125571469 |
- |
- |
PREDICTED: S-type anion channel SLAH4-like [Jatropha curcas] |
| 19 |
Hb_170597_010 |
0.1127696877 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 20 |
Hb_004137_090 |
0.1151948707 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |