| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
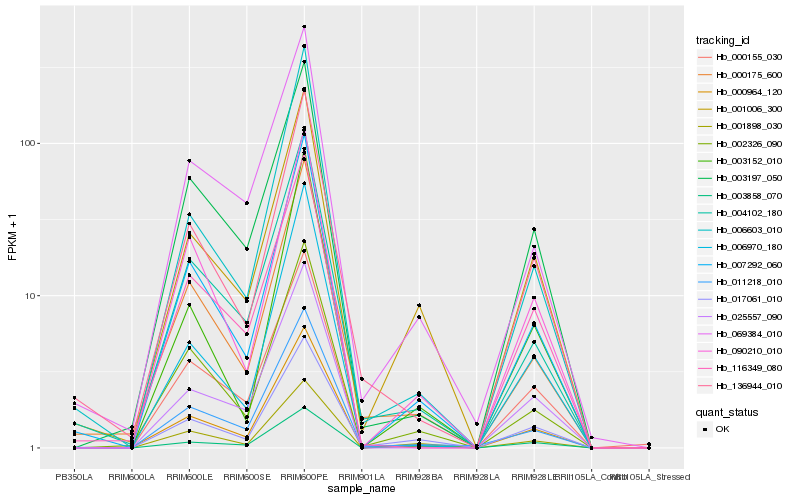
Hb_000964_120 |
0.0 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 2 |
Hb_007292_060 |
0.0573417555 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 3 |
Hb_002326_090 |
0.0701513213 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_001006_300 |
0.0775298142 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 5 |
Hb_003197_050 |
0.079211356 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 6 |
Hb_011218_010 |
0.0799598 |
- |
- |
PREDICTED: putative indole-3-acetic acid-amido synthetase GH3.9 [Jatropha curcas] |
| 7 |
Hb_090210_010 |
0.080378274 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 8 |
Hb_000155_030 |
0.0827063102 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 9 |
Hb_017061_010 |
0.082780351 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 10 |
Hb_000175_600 |
0.0851488956 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 11 |
Hb_025557_090 |
0.0859414289 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 12 |
Hb_001898_030 |
0.08674134 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 13 |
Hb_116349_080 |
0.0867743174 |
- |
- |
Nectarin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_004102_180 |
0.0882662109 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_136944_010 |
0.0903610709 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 16 |
Hb_069384_010 |
0.0904582578 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003858_070 |
0.0942289518 |
- |
- |
PREDICTED: uncharacterized protein LOC105647879 [Jatropha curcas] |
| 18 |
Hb_006970_180 |
0.0955320486 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 19 |
Hb_006603_010 |
0.0960178656 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 20 |
Hb_003152_010 |
0.0962992439 |
- |
- |
- |