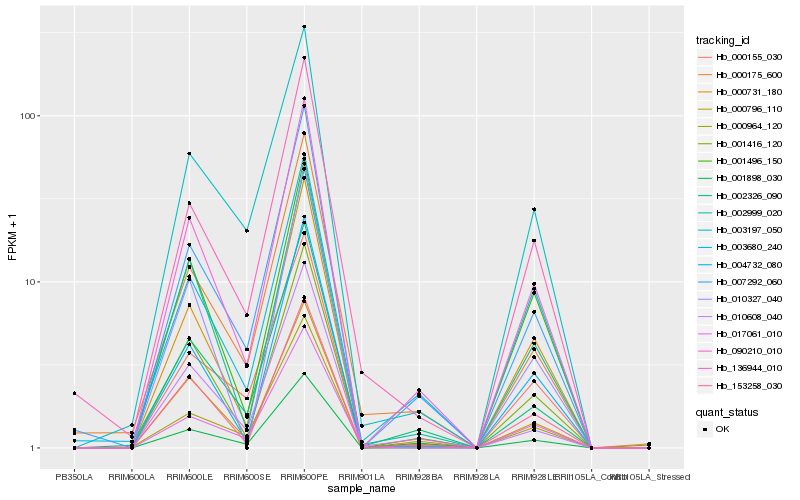
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_090210_010 |
0.0 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 2 |
Hb_153258_030 |
0.055379854 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 3 |
Hb_000731_180 |
0.0597488238 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 4 |
Hb_000796_110 |
0.0631848618 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 5 |
Hb_007292_060 |
0.0632214464 |
- |
- |
PREDICTED: uncharacterized protein LOC105632853 [Jatropha curcas] |
| 6 |
Hb_002326_090 |
0.0717043747 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_002999_020 |
0.079596809 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 8 |
Hb_001416_120 |
0.0799638889 |
- |
- |
caspase, putative [Ricinus communis] |
| 9 |
Hb_001898_030 |
0.0802528748 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |
| 10 |
Hb_000964_120 |
0.080378274 |
- |
- |
DNA-damage-inducible protein f, putative [Ricinus communis] |
| 11 |
Hb_004732_080 |
0.081678983 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 12 |
Hb_003197_050 |
0.0938696011 |
- |
- |
hypothetical protein JCGZ_04279 [Jatropha curcas] |
| 13 |
Hb_001496_150 |
0.0939758337 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 14 |
Hb_003680_240 |
0.0965041969 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 15 |
Hb_000175_600 |
0.0989745287 |
- |
- |
PREDICTED: TATA-binding protein-associated factor 2N isoform X2 [Jatropha curcas] |
| 16 |
Hb_136944_010 |
0.0998497855 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 17 |
Hb_017061_010 |
0.1000967954 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 18 |
Hb_000155_030 |
0.1006827435 |
- |
- |
PREDICTED: uncharacterized protein LOC105649591 [Jatropha curcas] |
| 19 |
Hb_010608_040 |
0.1053763702 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 20 |
Hb_010327_040 |
0.1068545483 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |