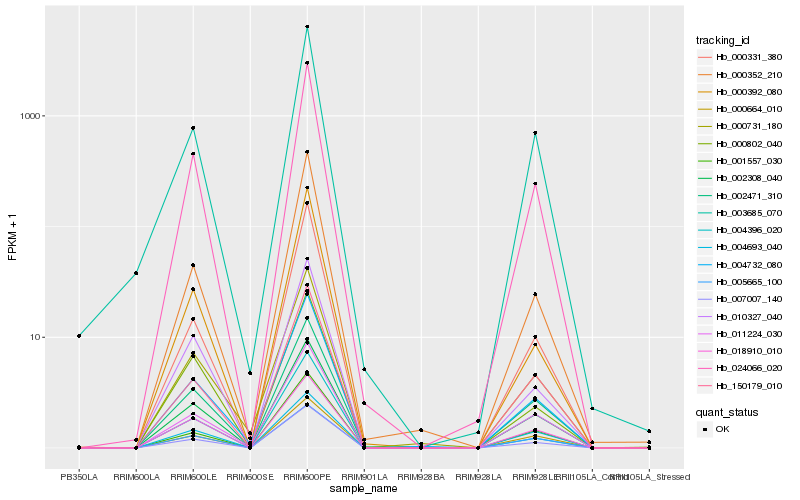
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024066_020 |
0.0 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 2 |
Hb_007007_140 |
0.0216658177 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 3 |
Hb_004396_020 |
0.0326503134 |
- |
- |
SAUR-like auxin-responsive protein family [Theobroma cacao] |
| 4 |
Hb_004732_080 |
0.0501647154 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Populus euphratica] |
| 5 |
Hb_002308_040 |
0.0531518698 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 6 |
Hb_004693_040 |
0.0547614885 |
- |
- |
- |
| 7 |
Hb_002471_310 |
0.0559855084 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_010327_040 |
0.0593423263 |
- |
- |
hypothetical protein POPTR_0004s17300g [Populus trichocarpa] |
| 9 |
Hb_000731_180 |
0.0683031838 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 10 |
Hb_150179_010 |
0.0694780112 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |
| 11 |
Hb_011224_030 |
0.0703263845 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_003685_070 |
0.0725493095 |
- |
- |
RecName: Full=(S)-hydroxynitrile lyase; AltName: Full=(S)-acetone-cyanohydrin lyase; AltName: Full=Oxynitrilase [Hevea brasiliensis] |
| 13 |
Hb_000802_040 |
0.0798621221 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 14 |
Hb_000352_210 |
0.0805535283 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 15 |
Hb_000664_010 |
0.0805590028 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 16 |
Hb_000331_380 |
0.0806951685 |
- |
- |
hypothetical protein POPTR_0012s09560g [Populus trichocarpa] |
| 17 |
Hb_000392_080 |
0.0809611501 |
- |
- |
hypothetical protein glysoja_034865 [Glycine soja] |
| 18 |
Hb_001557_030 |
0.0816772883 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 19 |
Hb_018910_010 |
0.0827811756 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 20 |
Hb_005665_100 |
0.083716842 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |