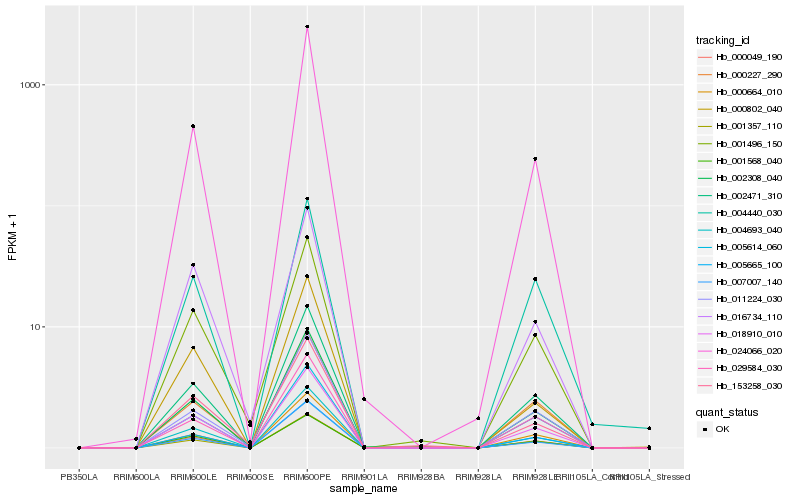
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018910_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 2 |
Hb_000049_190 |
0.0253525038 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 3 |
Hb_004693_040 |
0.0320742865 |
- |
- |
- |
| 4 |
Hb_001568_040 |
0.0364657858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005665_100 |
0.0377139898 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 6 |
Hb_002308_040 |
0.051234598 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 7 |
Hb_002471_310 |
0.0561144322 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005614_060 |
0.0678626236 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 9 |
Hb_001496_150 |
0.0686002441 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 10 |
Hb_000664_010 |
0.0688806098 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 11 |
Hb_001357_110 |
0.0786499586 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 12 |
Hb_016734_110 |
0.0805642578 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 13 |
Hb_029584_030 |
0.0826627836 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 14 |
Hb_024066_020 |
0.0827811756 |
- |
- |
P66 protein [Hevea brasiliensis] |
| 15 |
Hb_000227_290 |
0.0861576655 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 16 |
Hb_007007_140 |
0.086853342 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL6 [Jatropha curcas] |
| 17 |
Hb_153258_030 |
0.0882790214 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 18 |
Hb_004440_030 |
0.0891209031 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 19 |
Hb_011224_030 |
0.0921048237 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000802_040 |
0.0923501848 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |