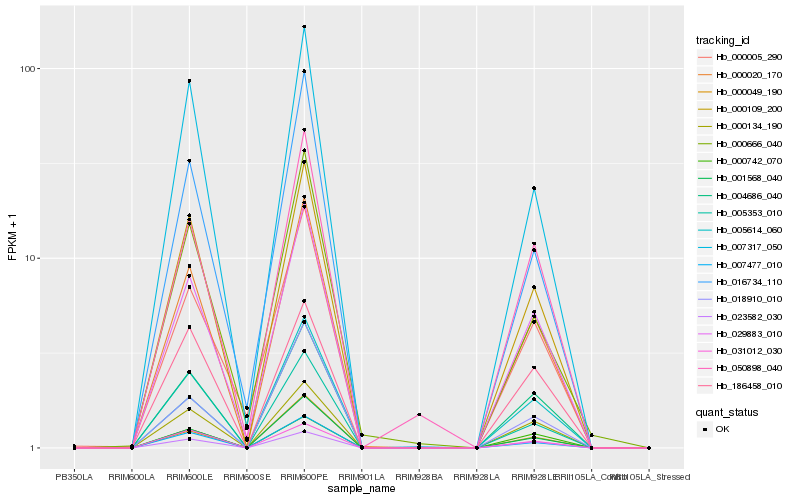
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_170 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 2 |
Hb_004686_040 |
0.0438388345 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |
| 3 |
Hb_000109_200 |
0.0470458882 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 4 |
Hb_029883_010 |
0.051229593 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 5 |
Hb_001568_040 |
0.0644125142 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000134_190 |
0.0705957896 |
- |
- |
hypothetical protein JCGZ_16725 [Jatropha curcas] |
| 7 |
Hb_007317_050 |
0.077756905 |
- |
- |
hypothetical protein POPTR_0009s16550g [Populus trichocarpa] |
| 8 |
Hb_000049_190 |
0.0816713588 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 9 |
Hb_050898_040 |
0.0853991489 |
- |
- |
- |
| 10 |
Hb_023582_030 |
0.0911221878 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_016734_110 |
0.0938452093 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 12 |
Hb_018910_010 |
0.0999690705 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X3 [Citrus sinensis] |
| 13 |
Hb_000005_290 |
0.1025214161 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 14 |
Hb_000666_040 |
0.1045406387 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 15 |
Hb_007477_010 |
0.1056380407 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 16 |
Hb_005614_060 |
0.1060586409 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 17 |
Hb_031012_030 |
0.1073896985 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 18 |
Hb_186458_010 |
0.1105181963 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 19 |
Hb_005353_010 |
0.1122380981 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000742_070 |
0.1122384746 |
- |
- |
PREDICTED: monoglyceride lipase isoform X2 [Bubalus bubalis] |