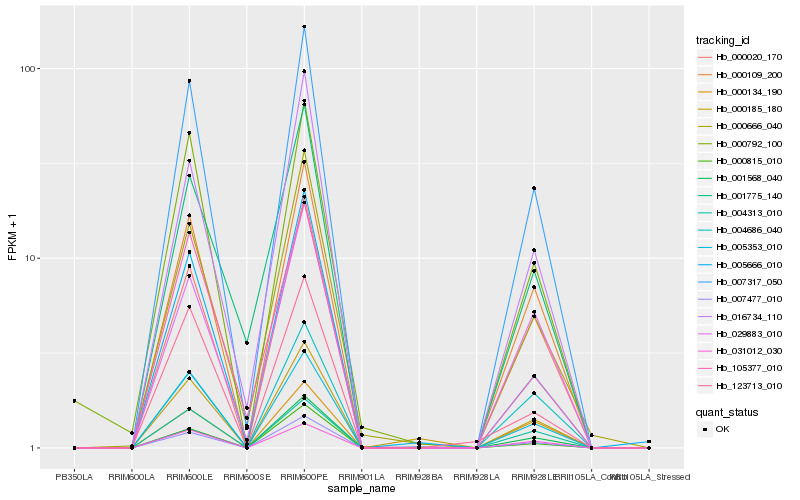
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007317_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s16550g [Populus trichocarpa] |
| 2 |
Hb_000109_200 |
0.050232303 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 3 |
Hb_005353_010 |
0.0567968806 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000020_170 |
0.077756905 |
- |
- |
PREDICTED: aspartic proteinase PCS1 [Jatropha curcas] |
| 5 |
Hb_016734_110 |
0.0879380843 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 6 |
Hb_000666_040 |
0.0886323449 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 7 |
Hb_031012_030 |
0.0890271544 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 8 |
Hb_005666_010 |
0.0892092155 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 9 |
Hb_000792_100 |
0.0947843986 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 10 |
Hb_029883_010 |
0.1000858521 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 11 |
Hb_007477_010 |
0.1005012473 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 12 |
Hb_123713_010 |
0.1013230102 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_004686_040 |
0.1051770826 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |
| 14 |
Hb_000815_010 |
0.1065294972 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105117338 [Populus euphratica] |
| 15 |
Hb_004313_010 |
0.1067673234 |
- |
- |
PREDICTED: patatin-like protein 1 [Prunus mume] |
| 16 |
Hb_001568_040 |
0.1100794722 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001775_140 |
0.1103547439 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 18 |
Hb_105377_010 |
0.1116877367 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 19 |
Hb_000134_190 |
0.1175122333 |
- |
- |
hypothetical protein JCGZ_16725 [Jatropha curcas] |
| 20 |
Hb_000185_180 |
0.1197761383 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |