| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
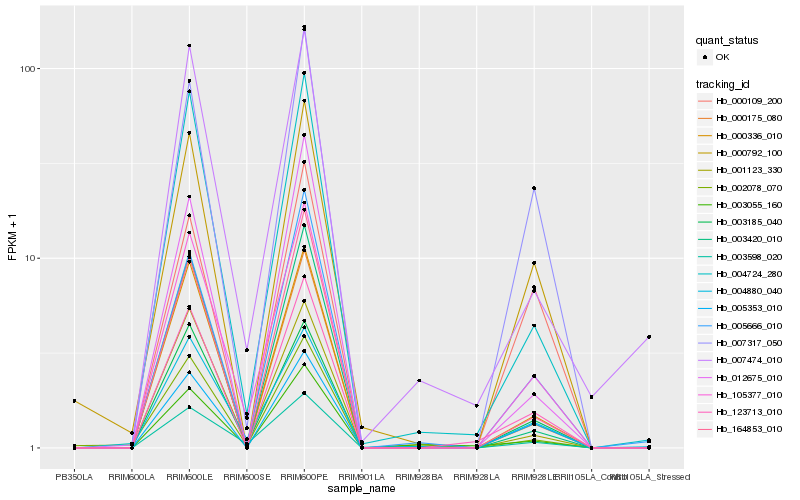
Hb_123713_010 |
0.0 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 2 |
Hb_005353_010 |
0.0898002418 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000336_010 |
0.0910511502 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 4 |
Hb_164853_010 |
0.0972022976 |
- |
- |
Patatin precursor, putative [Ricinus communis] |
| 5 |
Hb_004724_280 |
0.0985952849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_105377_010 |
0.1004894663 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 7 |
Hb_007317_050 |
0.1013230102 |
- |
- |
hypothetical protein POPTR_0009s16550g [Populus trichocarpa] |
| 8 |
Hb_003055_160 |
0.1013904985 |
- |
- |
PREDICTED: S-linalool synthase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000792_100 |
0.1049779452 |
- |
- |
PREDICTED: uncharacterized protein OsI_027940-like [Jatropha curcas] |
| 10 |
Hb_005666_010 |
0.1081146527 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 11 |
Hb_003420_010 |
0.111202304 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000175_080 |
0.1142486471 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 13 |
Hb_003185_040 |
0.1157266989 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 14 |
Hb_012675_010 |
0.1182844984 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_002078_070 |
0.1268732892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001123_330 |
0.1289850257 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 17 |
Hb_000109_200 |
0.1333214158 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 18 |
Hb_004880_040 |
0.1358590939 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003598_020 |
0.1362542205 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 20 |
Hb_007474_010 |
0.1379707599 |
- |
- |
vacuolar invertase [Manihot esculenta] |