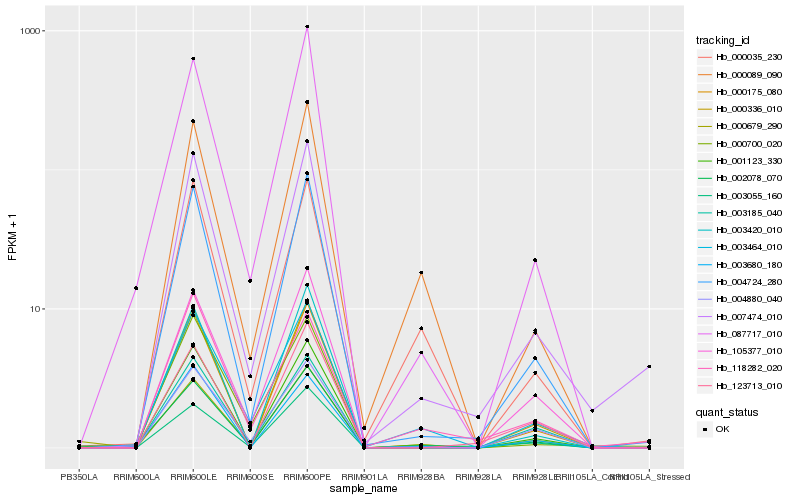
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007474_010 |
0.0 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 2 |
Hb_004724_280 |
0.0802361212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001123_330 |
0.0900105119 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 4 |
Hb_000336_010 |
0.1088837647 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 5 |
Hb_004880_040 |
0.1132651245 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000175_080 |
0.1177920592 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 7 |
Hb_003464_010 |
0.1184159052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003055_160 |
0.1190079885 |
- |
- |
PREDICTED: S-linalool synthase isoform X1 [Jatropha curcas] |
| 9 |
Hb_003185_040 |
0.1203326811 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 10 |
Hb_105377_010 |
0.1219235374 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 11 |
Hb_000700_020 |
0.1238495446 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 12 |
Hb_003420_010 |
0.1262746078 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_087717_010 |
0.1268171682 |
- |
- |
hypothetical protein JCGZ_01246 [Jatropha curcas] |
| 14 |
Hb_002078_070 |
0.1275196096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000089_090 |
0.1309132533 |
- |
- |
hypothetical protein POPTR_0019s12670g [Populus trichocarpa] |
| 16 |
Hb_000035_230 |
0.1332493209 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 17 |
Hb_118282_020 |
0.1362561826 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 18 |
Hb_003680_180 |
0.1372632582 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 19 |
Hb_123713_010 |
0.1379707599 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 20 |
Hb_000679_290 |
0.1399141408 |
transcription factor |
TF Family: AUX/IAA |
ATP binding protein, putative [Ricinus communis] |