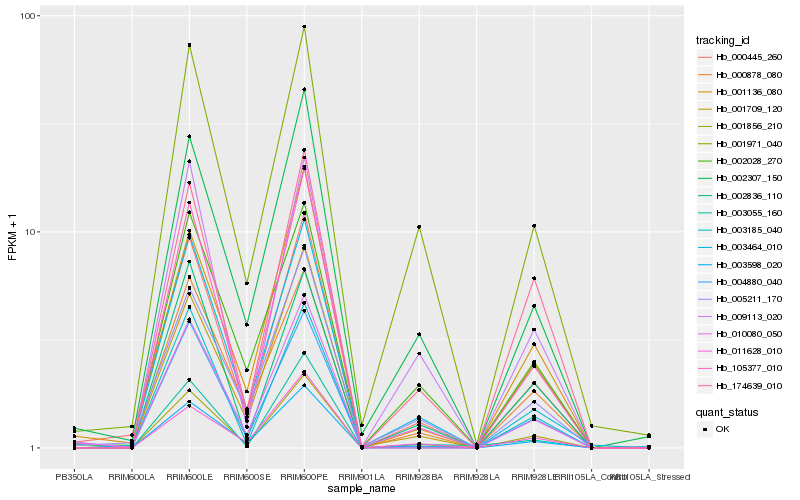
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001856_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 2 |
Hb_000445_260 |
0.0778308773 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 3 |
Hb_174639_010 |
0.0785799047 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_010080_050 |
0.08431547 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 5 |
Hb_000878_080 |
0.0853203533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 6 |
Hb_004880_040 |
0.0892860868 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |
| 7 |
Hb_009113_020 |
0.0912863415 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 8 |
Hb_003464_010 |
0.0990821441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005211_170 |
0.0991253632 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 10 |
Hb_003185_040 |
0.0991501997 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 11 |
Hb_011628_010 |
0.0999373233 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 12 |
Hb_002307_150 |
0.1011139324 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 13 |
Hb_001709_120 |
0.1029316712 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 14 |
Hb_105377_010 |
0.1032247436 |
- |
- |
hypothetical protein PRUPE_ppa001414mg [Prunus persica] |
| 15 |
Hb_002836_110 |
0.1047704347 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 16 |
Hb_003055_160 |
0.1084327291 |
- |
- |
PREDICTED: S-linalool synthase isoform X1 [Jatropha curcas] |
| 17 |
Hb_001971_040 |
0.1095205667 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 18 |
Hb_001136_080 |
0.1103531548 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 19 |
Hb_003598_020 |
0.1128534571 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 20 |
Hb_002028_270 |
0.1136149226 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |