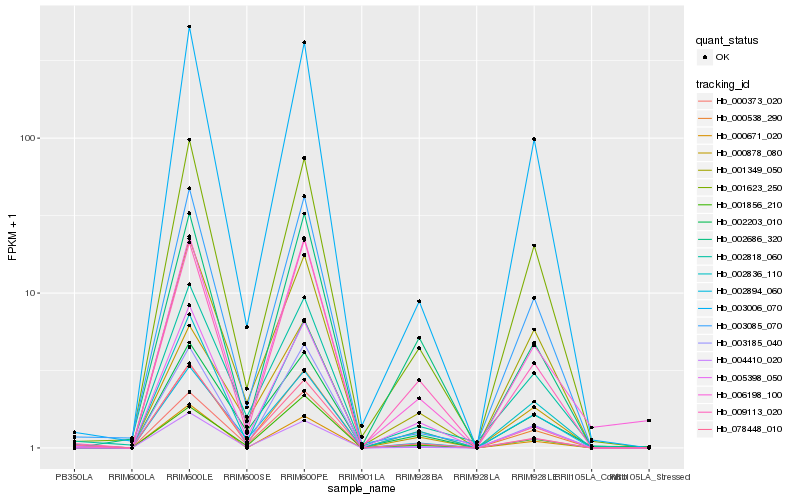
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002836_110 |
0.0 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 2 |
Hb_009113_020 |
0.0643843249 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 3 |
Hb_001623_250 |
0.0695445897 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 4 |
Hb_003006_070 |
0.0750734481 |
- |
- |
aquaporin [Manihot esculenta] |
| 5 |
Hb_001349_050 |
0.0891906661 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 6 |
Hb_003085_070 |
0.0900035673 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000538_290 |
0.0984909277 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 8 |
Hb_000373_020 |
0.1003534408 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 9 |
Hb_004410_020 |
0.1017397502 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 10 |
Hb_002818_060 |
0.1029825397 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_003185_040 |
0.1039713698 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 12 |
Hb_001856_210 |
0.1047704347 |
- |
- |
PREDICTED: uncharacterized protein LOC105644171 [Jatropha curcas] |
| 13 |
Hb_000671_020 |
0.1051210135 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |
| 14 |
Hb_006198_100 |
0.1052807659 |
- |
- |
PREDICTED: RNA-binding protein 24-B isoform X1 [Jatropha curcas] |
| 15 |
Hb_002686_320 |
0.106228044 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_002894_060 |
0.1069600527 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 17 |
Hb_005398_050 |
0.1079228933 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 18 |
Hb_078448_010 |
0.1094864731 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 19 |
Hb_000878_080 |
0.1155412678 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 20 |
Hb_002203_010 |
0.1158758064 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |