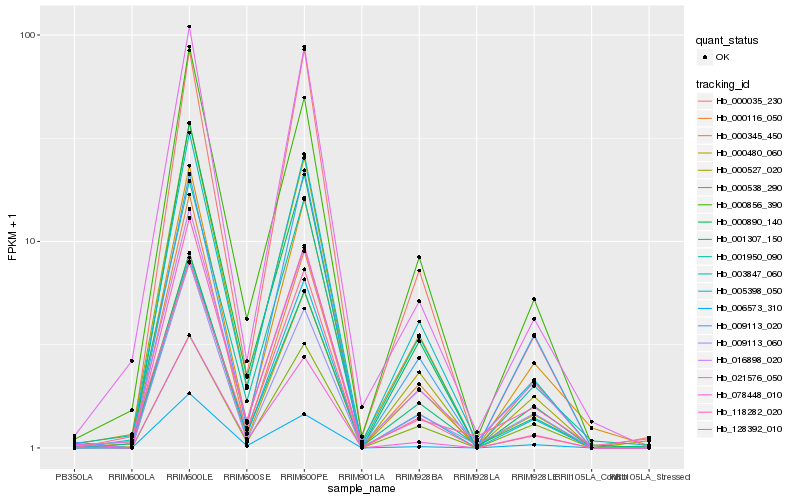
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005398_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003847_060 |
0.0658625071 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 3 |
Hb_000527_020 |
0.0776871625 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 4 |
Hb_078448_010 |
0.0805019877 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 5 |
Hb_000035_230 |
0.0824384321 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 6 |
Hb_001307_150 |
0.0868686548 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_016898_020 |
0.0877207486 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 8 |
Hb_009113_060 |
0.0893853394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000890_140 |
0.0900786113 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 10 |
Hb_000856_390 |
0.0920021435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001950_090 |
0.0924540371 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1C [Jatropha curcas] |
| 12 |
Hb_000538_290 |
0.0932839512 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 13 |
Hb_000116_050 |
0.0935082156 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_118282_020 |
0.09439437 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 15 |
Hb_009113_020 |
0.0947804697 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 16 |
Hb_000480_060 |
0.0950165978 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 17 |
Hb_021576_050 |
0.0971990038 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000345_450 |
0.0972613963 |
- |
- |
PREDICTED: microtubule-binding protein TANGLED [Jatropha curcas] |
| 19 |
Hb_128392_010 |
0.0990219679 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 20 |
Hb_006573_310 |
0.0996798354 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |