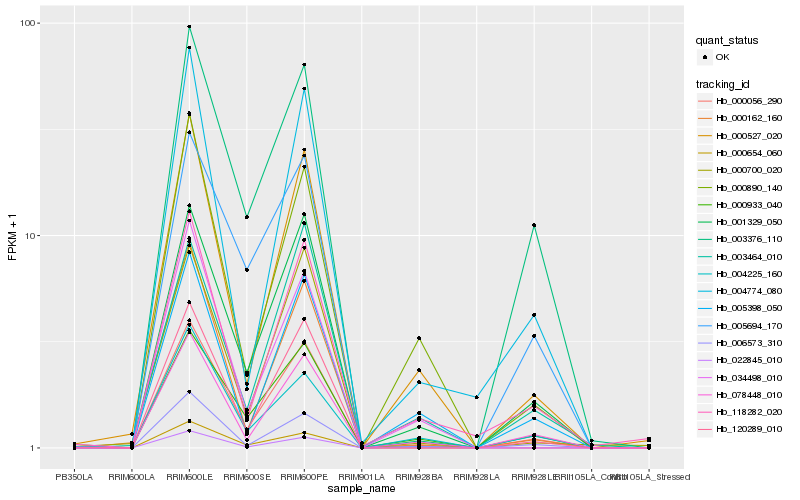
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_290 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001329_050 |
0.0546167554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_120289_010 |
0.0703754934 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 4 |
Hb_006573_310 |
0.0883439015 |
- |
- |
Tellurite resistance protein tehA, putative [Ricinus communis] |
| 5 |
Hb_000933_040 |
0.0915739185 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 6 |
Hb_078448_010 |
0.1023877305 |
- |
- |
hypothetical protein L484_017526 [Morus notabilis] |
| 7 |
Hb_000527_020 |
0.1031892915 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 8 |
Hb_000162_160 |
0.1047023342 |
- |
- |
PREDICTED: auxin transporter-like protein 5 [Populus euphratica] |
| 9 |
Hb_003376_110 |
0.1097002498 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000700_020 |
0.1136064521 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 11 |
Hb_118282_020 |
0.1161284022 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 12 |
Hb_022845_010 |
0.1176643019 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 13 |
Hb_004774_080 |
0.1178634438 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005694_170 |
0.1212572325 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005398_050 |
0.1243775414 |
- |
- |
PREDICTED: uncharacterized protein LOC105644215 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004225_160 |
0.1245092354 |
- |
- |
PREDICTED: kiwellin-like [Jatropha curcas] |
| 17 |
Hb_003464_010 |
0.1292839998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000654_060 |
0.1303348458 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 19 |
Hb_000890_140 |
0.1303740374 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 20 |
Hb_034498_010 |
0.1312992771 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |