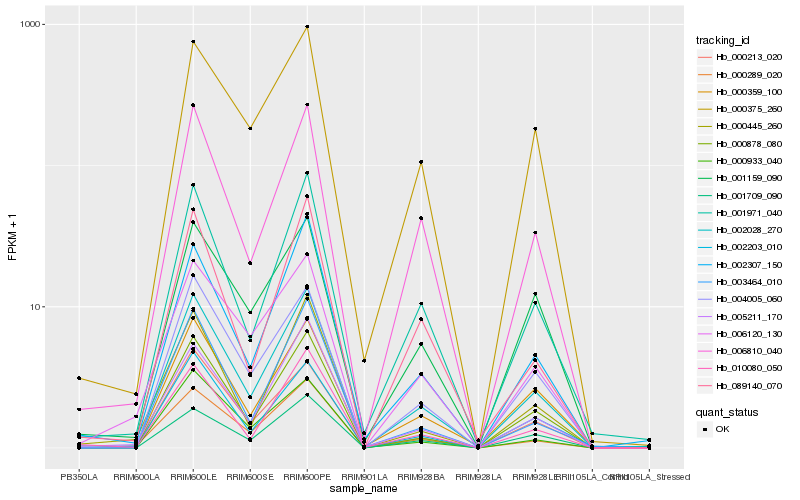
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_270 |
0.0 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001971_040 |
0.0666732646 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 3 |
Hb_006810_040 |
0.0751701639 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000878_080 |
0.0753303082 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 5 |
Hb_000359_100 |
0.0822237863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004005_060 |
0.0868004426 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 7 |
Hb_000289_020 |
0.0894422754 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 8 |
Hb_005211_170 |
0.090576244 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 9 |
Hb_002307_150 |
0.0944340846 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 10 |
Hb_010080_050 |
0.0951042765 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 11 |
Hb_002203_010 |
0.0957480154 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 12 |
Hb_001709_090 |
0.0961814044 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 13 |
Hb_000375_260 |
0.0972147131 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 14 |
Hb_000213_020 |
0.1006832101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003464_010 |
0.1029324808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_089140_070 |
0.1064469344 |
- |
- |
PREDICTED: uncharacterized protein LOC105637961 [Jatropha curcas] |
| 17 |
Hb_000445_260 |
0.1104349021 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 18 |
Hb_001159_090 |
0.1106144838 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 19 |
Hb_006120_130 |
0.1113200065 |
- |
- |
- |
| 20 |
Hb_000933_040 |
0.1120409202 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |