| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
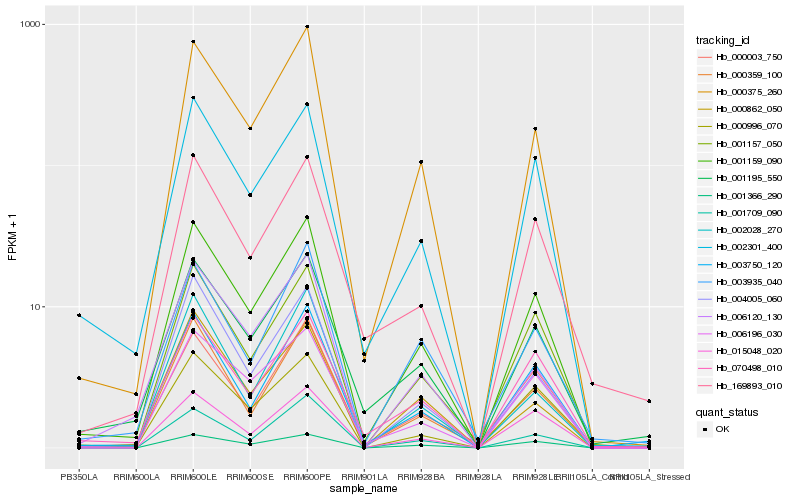
Hb_001159_090 |
0.0 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 2 |
Hb_000375_260 |
0.0543368659 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 3 |
Hb_002301_400 |
0.0819837205 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 4 |
Hb_001157_050 |
0.0835797116 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 5 |
Hb_006196_030 |
0.0845196837 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 6 |
Hb_003750_120 |
0.0865238577 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 7 |
Hb_000359_100 |
0.0949526341 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001195_550 |
0.0955343693 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001366_290 |
0.0967816308 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_000003_750 |
0.0969285301 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 11 |
Hb_004005_060 |
0.0996023957 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 12 |
Hb_001709_090 |
0.1039092817 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 13 |
Hb_000996_070 |
0.1048634219 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_070498_010 |
0.1059719068 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 15 |
Hb_000862_050 |
0.1104890821 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 16 |
Hb_003935_040 |
0.1105662593 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 17 |
Hb_002028_270 |
0.1106144838 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 18 |
Hb_006120_130 |
0.1113585034 |
- |
- |
- |
| 19 |
Hb_169893_010 |
0.1118679877 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 20 |
Hb_015048_020 |
0.1123100692 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |