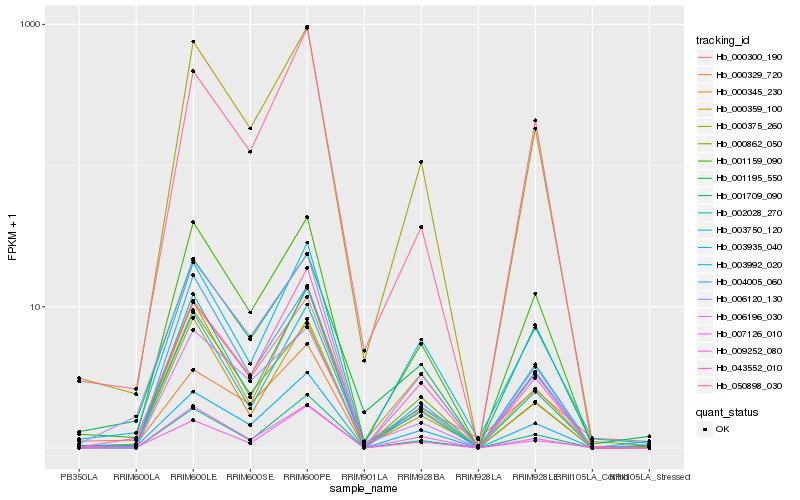
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000375_260 |
0.0 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K4 [Manihot esculenta] |
| 2 |
Hb_001159_090 |
0.0543368659 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 3 |
Hb_001709_090 |
0.0811265609 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 4 |
Hb_043552_010 |
0.0908294294 |
- |
- |
PREDICTED: pectinesterase/pectinesterase inhibitor PPE8B isoform X1 [Jatropha curcas] |
| 5 |
Hb_006120_130 |
0.0961968392 |
- |
- |
- |
| 6 |
Hb_002028_270 |
0.0972147131 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 7 |
Hb_003992_020 |
0.1018630367 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000359_100 |
0.1046765574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004005_060 |
0.1059286579 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 10 |
Hb_000345_230 |
0.1072156973 |
- |
- |
- |
| 11 |
Hb_003935_040 |
0.1075600109 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP [Jatropha curcas] |
| 12 |
Hb_009252_080 |
0.1085111556 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 13 |
Hb_006196_030 |
0.1085479444 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 14 |
Hb_000862_050 |
0.1108384924 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 15 |
Hb_001195_550 |
0.1137721256 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003750_120 |
0.1150550565 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 17 |
Hb_000329_720 |
0.1169056509 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 18 |
Hb_000300_190 |
0.1177645141 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 19 |
Hb_007126_010 |
0.1178993128 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Jatropha curcas] |
| 20 |
Hb_050898_030 |
0.1182108416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |