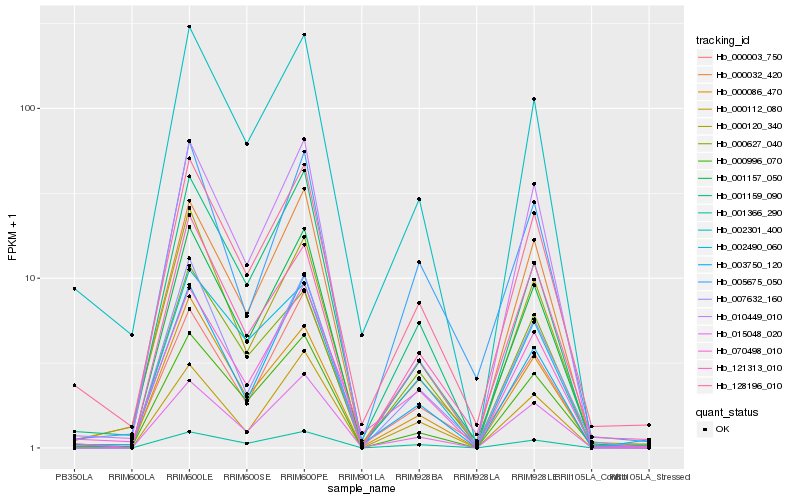
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001157_050 |
0.0 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 2 |
Hb_015048_020 |
0.0700669577 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 3 |
Hb_007632_160 |
0.0711025517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001366_290 |
0.075040058 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_000003_750 |
0.0772291456 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 6 |
Hb_000996_070 |
0.0775351705 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_128196_010 |
0.0830359453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001159_090 |
0.0835797116 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 9 |
Hb_000032_420 |
0.0895794052 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 10 |
Hb_000086_470 |
0.0917916739 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 11 |
Hb_070498_010 |
0.0941001267 |
- |
- |
receptor kinase, putative [Ricinus communis] |
| 12 |
Hb_002301_400 |
0.0951243749 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 13 |
Hb_000112_080 |
0.0954852319 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 14 |
Hb_000120_340 |
0.0954896674 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 15 |
Hb_010449_010 |
0.0957212443 |
- |
- |
l-ascorbate oxidase, putative [Ricinus communis] |
| 16 |
Hb_000627_040 |
0.0965346309 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 17 |
Hb_005675_050 |
0.0983896358 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 18 |
Hb_121313_010 |
0.099580376 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 19 |
Hb_003750_120 |
0.0997431819 |
- |
- |
PREDICTED: uncharacterized protein LOC105112992 [Populus euphratica] |
| 20 |
Hb_002490_060 |
0.1006948152 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |