| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
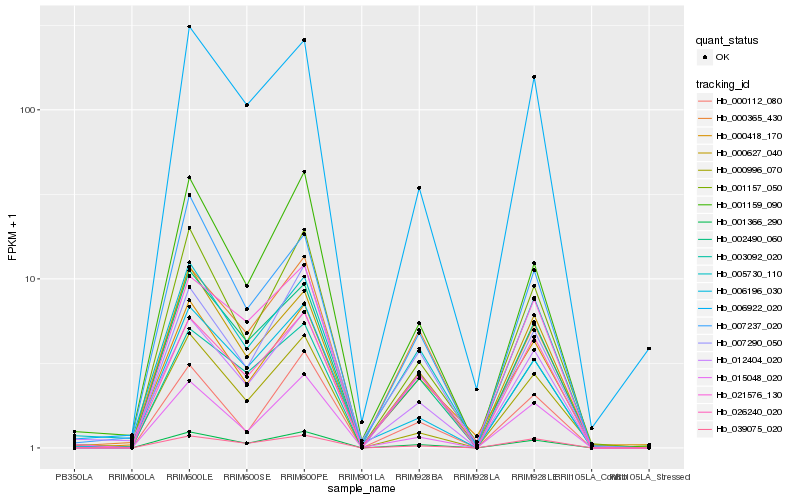
Hb_001366_290 |
0.0 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_002490_060 |
0.0692216747 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 3 |
Hb_001157_050 |
0.075040058 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 4 |
Hb_039075_020 |
0.0879671173 |
- |
- |
PREDICTED: uncharacterized protein LOC103331245 [Prunus mume] |
| 5 |
Hb_000627_040 |
0.0888646923 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 6 |
Hb_015048_020 |
0.0905289055 |
- |
- |
PREDICTED: myb-like protein D [Jatropha curcas] |
| 7 |
Hb_007290_050 |
0.0931741788 |
- |
- |
PREDICTED: uncharacterized protein LOC105629472 isoform X1 [Jatropha curcas] |
| 8 |
Hb_026240_020 |
0.0957206634 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 9 |
Hb_012404_020 |
0.0959291986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001159_090 |
0.0967816308 |
- |
- |
PREDICTED: methylsterol monooxygenase 1-1-like [Jatropha curcas] |
| 11 |
Hb_006922_020 |
0.0977561975 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 12 |
Hb_006196_030 |
0.0980888039 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 13 |
Hb_021576_130 |
0.1005256977 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000365_430 |
0.101244556 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 15 |
Hb_000112_080 |
0.1020131667 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 16 |
Hb_000418_170 |
0.1042371044 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 17 |
Hb_000996_070 |
0.1075105474 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_003092_020 |
0.1075743306 |
- |
- |
PREDICTED: heptahelical transmembrane protein 1 [Jatropha curcas] |
| 19 |
Hb_007237_020 |
0.1133037786 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 20 |
Hb_005730_110 |
0.1148293075 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |