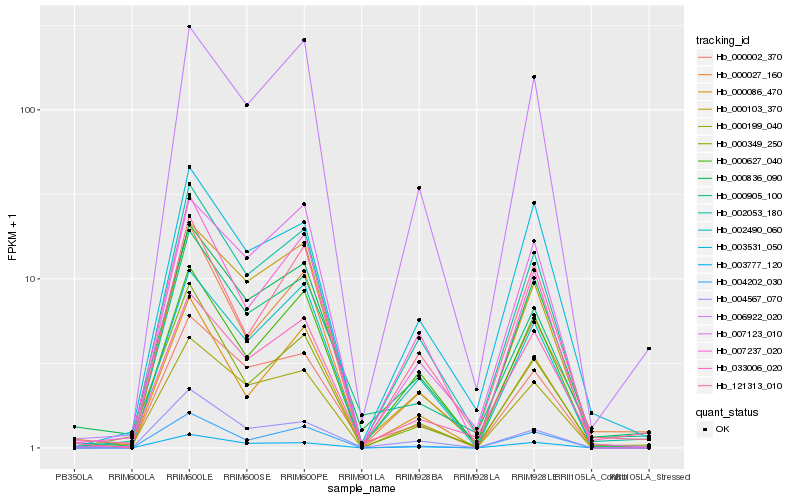
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002053_180 |
0.0 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 2 |
Hb_000199_040 |
0.0704409126 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000627_040 |
0.0802277132 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 4 |
Hb_007237_020 |
0.0817151722 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 5 |
Hb_006922_020 |
0.0841647844 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 6 |
Hb_000836_090 |
0.0914267468 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 7 |
Hb_002490_060 |
0.0934105519 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 8 |
Hb_003777_120 |
0.0952656511 |
- |
- |
- |
| 9 |
Hb_000002_370 |
0.0953035155 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 10 |
Hb_000027_160 |
0.0976027584 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 11 |
Hb_003531_050 |
0.0983244548 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_121313_010 |
0.0992006691 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 13 |
Hb_000086_470 |
0.0992507845 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 14 |
Hb_000349_250 |
0.1031180569 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 15 |
Hb_004202_030 |
0.1034782565 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 16 |
Hb_000103_370 |
0.103550559 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 17 |
Hb_000905_100 |
0.1084681334 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004567_070 |
0.1105515849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007123_010 |
0.111483425 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 20 |
Hb_033006_020 |
0.1170972947 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |