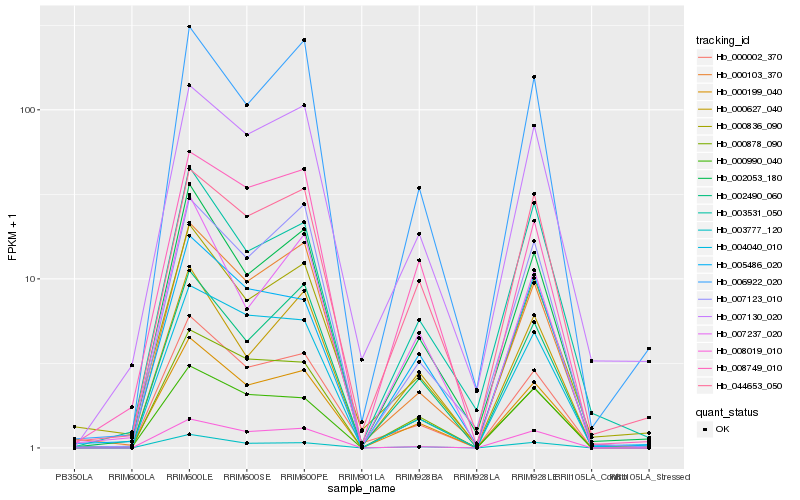
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000199_040 |
0.0 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002053_180 |
0.0704409126 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 3 |
Hb_000002_370 |
0.0771105462 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 4 |
Hb_003777_120 |
0.0811619524 |
- |
- |
- |
| 5 |
Hb_006922_020 |
0.0984707011 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 6 |
Hb_000878_090 |
0.0996167284 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 7 |
Hb_002490_060 |
0.1004839154 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 8 |
Hb_005486_020 |
0.1004938386 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 9 |
Hb_000836_090 |
0.1034504822 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 10 |
Hb_008749_010 |
0.1041299357 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 11 |
Hb_004040_010 |
0.1041766831 |
- |
- |
hypothetical protein POPTR_0019s14250g [Populus trichocarpa] |
| 12 |
Hb_044653_050 |
0.1067252153 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000103_370 |
0.1067635968 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 14 |
Hb_007130_020 |
0.1101418324 |
- |
- |
- |
| 15 |
Hb_008019_010 |
0.1108094272 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 16 |
Hb_007237_020 |
0.1109834878 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 17 |
Hb_000627_040 |
0.1123577811 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 18 |
Hb_000990_040 |
0.1158922704 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 19 |
Hb_003531_050 |
0.1176951362 |
- |
- |
PREDICTED: pyridoxal reductase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007123_010 |
0.1193816112 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |