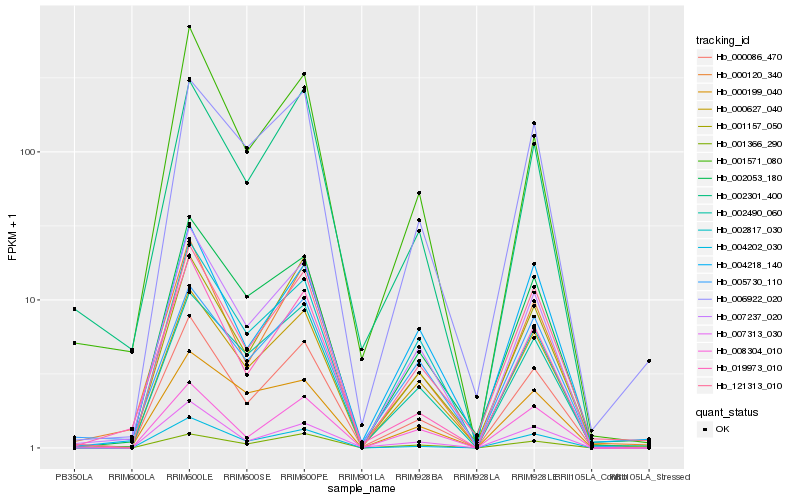
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007237_020 |
0.0 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 2 |
Hb_000086_470 |
0.0630450599 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 3 |
Hb_000627_040 |
0.0676722534 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 4 |
Hb_002053_180 |
0.0817151722 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_002490_060 |
0.0863610703 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 6 |
Hb_002817_030 |
0.0874918234 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 7 |
Hb_000120_340 |
0.0879463165 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 8 |
Hb_001571_080 |
0.0961189594 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 9 |
Hb_121313_010 |
0.100836282 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 10 |
Hb_007313_030 |
0.10142971 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 11 |
Hb_004202_030 |
0.1029151734 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 12 |
Hb_019973_010 |
0.1092147306 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000199_040 |
0.1109834878 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001157_050 |
0.1131873256 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 15 |
Hb_008304_010 |
0.113205967 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Fragaria vesca subsp. vesca] |
| 16 |
Hb_001366_290 |
0.1133037786 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_004218_140 |
0.1133171681 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |
| 18 |
Hb_006922_020 |
0.1139760088 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 19 |
Hb_002301_400 |
0.1140428598 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 20 |
Hb_005730_110 |
0.1144145807 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |