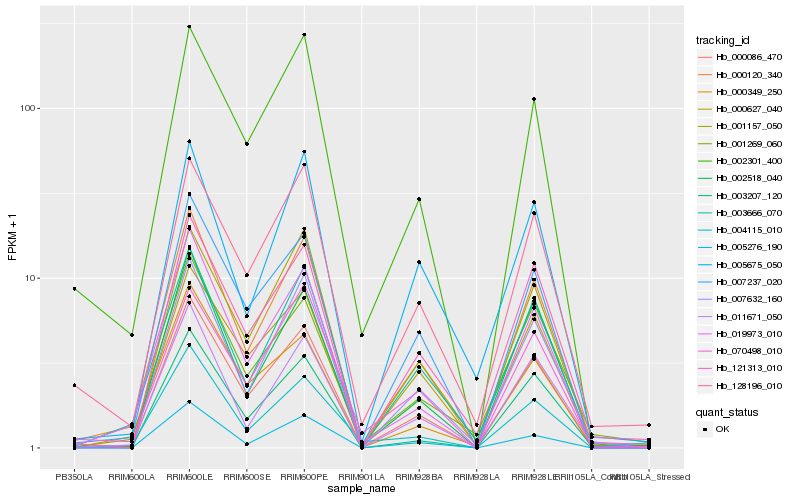
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_340 |
0.0 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 2 |
Hb_003666_070 |
0.0695598077 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 3 |
Hb_000086_470 |
0.0704013781 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 4 |
Hb_019973_010 |
0.0766117928 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 5 |
Hb_007632_160 |
0.0830024468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011671_050 |
0.08791752 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 7 |
Hb_007237_020 |
0.0879463165 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 8 |
Hb_001157_050 |
0.0954896674 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 9 |
Hb_004115_010 |
0.0959508673 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 10 |
Hb_121313_010 |
0.0971416778 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 11 |
Hb_002518_040 |
0.097774479 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 12 |
Hb_128196_010 |
0.0981881728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002301_400 |
0.1007622562 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 14 |
Hb_001269_060 |
0.105346563 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000627_040 |
0.1070648266 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 16 |
Hb_005675_050 |
0.1096060244 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 17 |
Hb_005276_190 |
0.1103163772 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003207_120 |
0.1103740179 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 19 |
Hb_000349_250 |
0.1116839322 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 20 |
Hb_070498_010 |
0.1124191083 |
- |
- |
receptor kinase, putative [Ricinus communis] |