| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
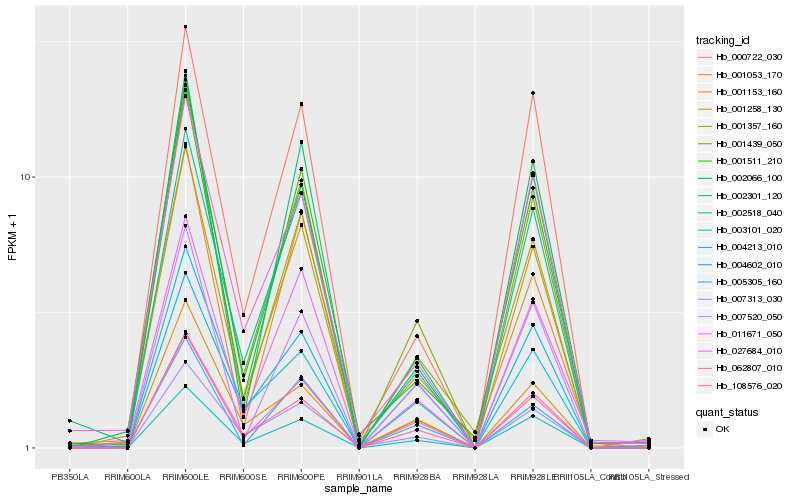
Hb_001439_050 |
0.0 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_003101_020 |
0.053113239 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 3 |
Hb_062807_010 |
0.0594030768 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 4 |
Hb_108576_020 |
0.0691711088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002518_040 |
0.0808019427 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 6 |
Hb_007313_030 |
0.0925112731 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_011671_050 |
0.0931591408 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 8 |
Hb_002066_100 |
0.0948360641 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 9 |
Hb_001153_160 |
0.0970725023 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 10 |
Hb_002301_120 |
0.0980903254 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 11 |
Hb_007520_050 |
0.0984612081 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 12 |
Hb_004213_010 |
0.0996931308 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_001258_130 |
0.1008084708 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 14 |
Hb_001511_210 |
0.1009712271 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 15 |
Hb_000722_030 |
0.1025419483 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 16 |
Hb_001053_170 |
0.1098603051 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 17 |
Hb_027684_010 |
0.1121723743 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 18 |
Hb_005305_160 |
0.1151803712 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001357_160 |
0.1162227331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004602_010 |
0.1169590405 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |