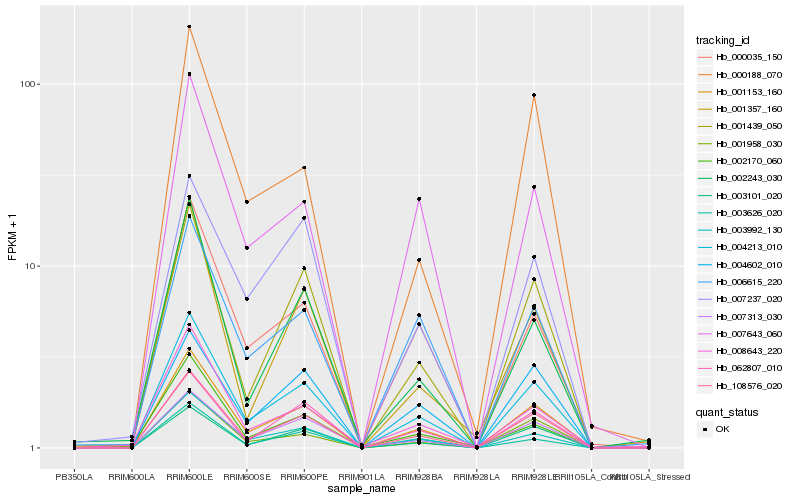
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004213_010 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001153_160 |
0.0052455288 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 3 |
Hb_108576_020 |
0.0363807772 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003992_130 |
0.0639802266 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000035_150 |
0.0924729601 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 6 |
Hb_007313_030 |
0.0952260468 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_003101_020 |
0.0991078863 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 8 |
Hb_001439_050 |
0.0996931308 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_062807_010 |
0.1052515123 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 10 |
Hb_001958_030 |
0.1083588032 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 11 |
Hb_007643_060 |
0.1085571285 |
- |
- |
- |
| 12 |
Hb_003626_020 |
0.1127963645 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 13 |
Hb_006615_220 |
0.1150699767 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_002170_060 |
0.1225600281 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 15 |
Hb_008643_220 |
0.123162152 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 16 |
Hb_004602_010 |
0.1259624877 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 17 |
Hb_002243_030 |
0.1276392977 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000188_070 |
0.1311989662 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 19 |
Hb_007237_020 |
0.1349207654 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 20 |
Hb_001357_160 |
0.1358188351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |