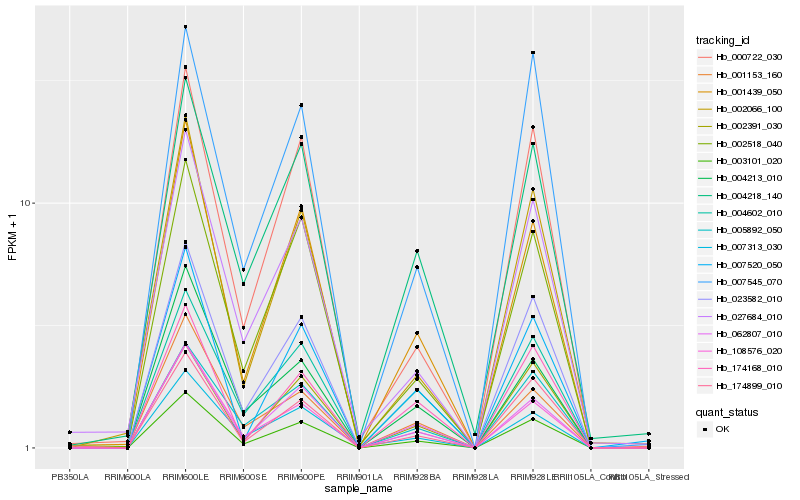
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003101_020 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 2 |
Hb_001439_050 |
0.053113239 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_108576_020 |
0.0650717293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_062807_010 |
0.0724342019 |
- |
- |
hypothetical protein JCGZ_15896 [Jatropha curcas] |
| 5 |
Hb_002518_040 |
0.075589492 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 6 |
Hb_002066_100 |
0.0850944401 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 7 |
Hb_000722_030 |
0.0852585404 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 8 |
Hb_005892_050 |
0.0869904799 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 9 |
Hb_007313_030 |
0.0875739967 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 10 |
Hb_174899_010 |
0.0909567564 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 11 |
Hb_004602_010 |
0.0923395717 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 12 |
Hb_023582_010 |
0.0924552358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001153_160 |
0.0958304164 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 14 |
Hb_007520_050 |
0.0981275091 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor WER-like [Jatropha curcas] |
| 15 |
Hb_004213_010 |
0.0991078863 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002391_030 |
0.0994642394 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |
| 17 |
Hb_007545_070 |
0.1011951123 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 18 |
Hb_027684_010 |
0.1013928009 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 19 |
Hb_174168_010 |
0.1025116927 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 20 |
Hb_004218_140 |
0.1040910241 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 isoform X3 [Jatropha curcas] |